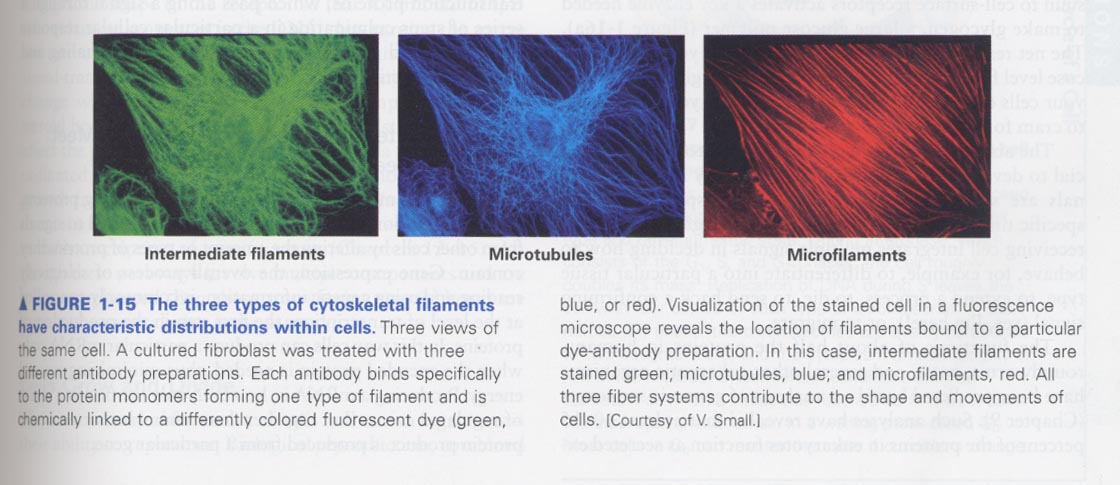
1장; 생물학의 각 특성분야가 생명체 (세포)의 이해에 어떠한 기여를 하고 있는 지 ?
(1) Cell Biology; size, shape, location
1. 1600년경 crude microscope
2. modern compound microscope; bacteria (1 μm) 측정가능, 0.2 μm사이의 물체구분
이용도--> 예) chromosome관찰; with a dye binding to DNA
target protein 관찰; with antibody linked to fluorescent dye (GFP)

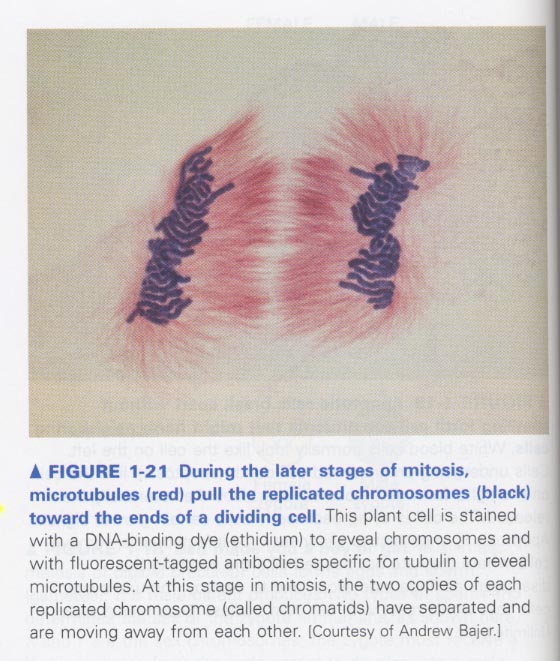
예) microtubules in the movement of chromosome; Ab against tubulin
3. Electron microscope
--> very thin sections, precluding examination of living cells, high resolution (0.1 nm)
(2) Biochemistry; structure and chemistry
1. fractionation
--> 물리, 화학적 특성에 따른 분리 예) 분자량, electric charge

2. antibody의 이용
--> for isolating larger amounts of a protein of interest
3. tag의 이용
--> to pull out the protein from whole cell extracts ex) 유전자에 붙임
4. 목적; a) how it catalyzes a chemical reaction and carry out other functions
b) how its activity is regulated
c) 기능과 연관된 strucure studies
(3) Genetics; damaged genes에 대한 분석--> 단백질에 대한 정확한 기능이해
1. mutation
** how can we isolate and maintain mutant organisms or cells ? --> temperature-sensitive mutants
--> 예) 세포분열 연관 유전자 발견
문제점; about which proteins they encode, how these proteins participate in the process
(4) Genomics; structure and expression of entire genomes
1. 이용도
--> evolutionary processes, in following the inheritance of diseases in human families
2. DNA microarray; gene expression변이 관찰 예) disease process, development, 어떤 신호자극시
 green
(no serum), red (serum)
green
(no serum), red (serum)
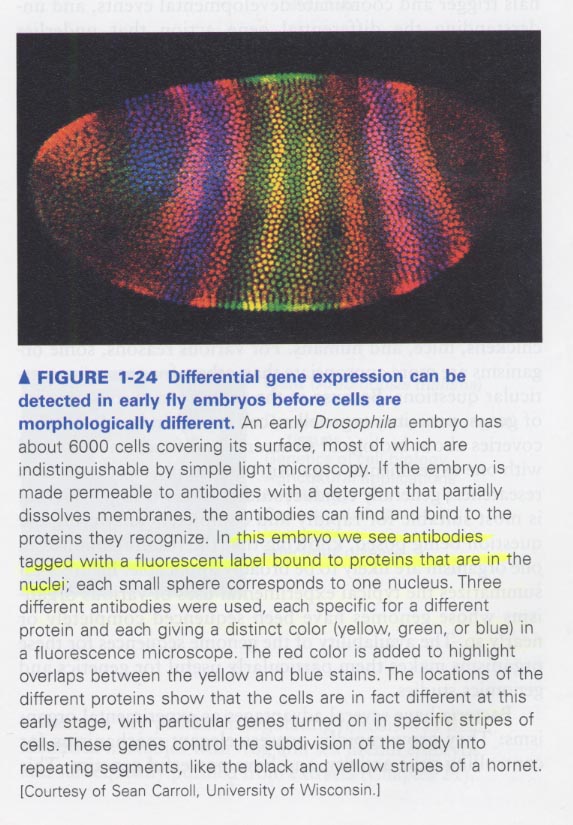
(5) Developmental biology
1. differnces among differentiated cells
--> specific sets of proteins의 발현차이 때문, 서로 다른 단백질을 blue, yellow, geen dye로 tagging
(6) 적절한 실험모델 설정
--> 조건; genomes have been sequenced or nearly so

(7) Plasmid Vectors
1. Bacterial plasmid
- double stranded circular DNA, 1 ~ 200 Kb
- can replicate and inherit independently of bacterial chromosome
depend on proteins and enzymes encoded by host
- own genes for enzymes which are advantageous to the host
2. Features
(1) replication
most cloning tasks
- in the relaxed vectors --> a greater yield of DNA/volume
- but, stringently controlled plasmids; use in cloning of lethal proteins which are deleterious to host
(2) mobilization
① in usual, bacterial conjugation transmit the plasmid
② vectors in common use;
⇒ lack of 'mob' gene -> no transmission by conjugation
but, a third plasmid(ColK) containing mob gene is present in cells -> possible to transmit
(3) selective markers
- after transformation, to identify the plasmids
- common use; (ex) Amp, Tet, Chloramphenicol, kanamycin
① Ampicillin
--> binds to many enzymes in the bacterial membrane; block the cell wall synthesis
--> AmpR gene encodes enzyme
transport to periplasmic space -> hydrolysis of beta-lactam ring -> detoxification of drug
② tetracycline
--> binds to 30S ribosomal subunit; inhibit the protein synthesis
--> TetR gene encodes a membrane-associated protein
prevent the antibiotic from entering into the cells
③ chloramphenicol
--> binds to 50S ribosomal subunit; inhibit the protein synthesis
--> cat (CmR) gene encodes a tetrameric cytosolic protein
catalyze the chloramphenicol; non-binding to 50S ribosomal subunit
(8) Gel electrophoresis
(종류) ① agarose ② polyacrylamide
-- 1-10ng DNA -- separation of small fragments (5-500 bp)
-- 0.2 - 50 kb -- 1 bp difference
(A) agarose gel electrophoresis
⇒ seaweed, a linear polymer
⇒ contaminated with polysaccharides, salts, proteins
⇒ low-melting gel (10-500 bp); usually high conc (4-10%), so purified DNA probably contaminated
** factors on movement
1) DNA size ; inversely proportional to log10bp (Fig 6-1)
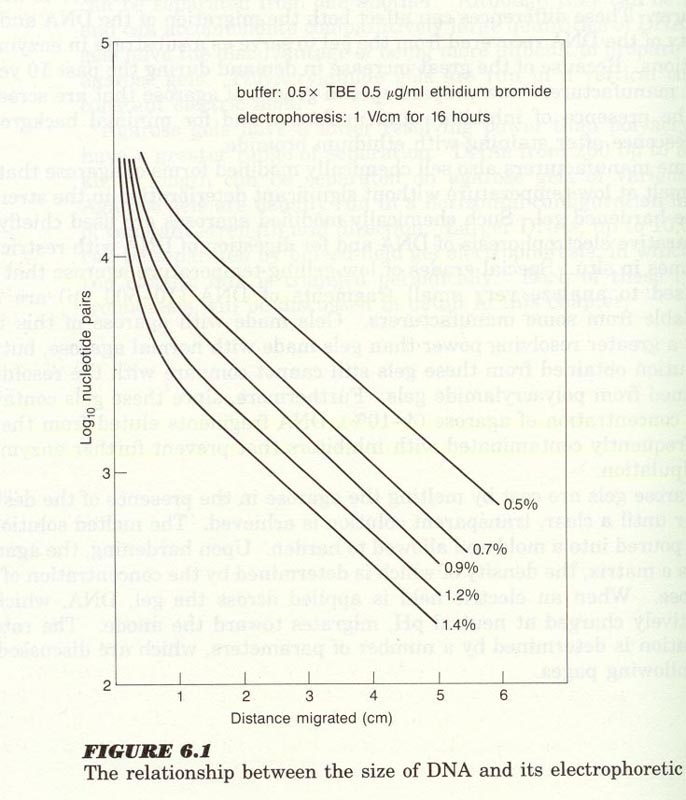
2) agarose conc.
3) DNA conformation ; supercoiled, nicked, linear
4) voltage
5) base composition and temp. ; no-effect (4oC-30oC)
(but low-melting gel run at 4oC)
6) EtBr ; ~ 15% reduced
7) buffer ; if H2O --> slow moving
if 10X buffer --> heat and gel melting
TAE : ⇒ buffering capacity is low and need change **
⇒ migration is fast (~10%)
⇒ supercoiled form resolution is good
TBE & TPE : more expensive
alkaline buffer (50mM NaOH/1mM EDTA): for ssDNA, first melt in water
(B) DNA recovery from agarose gels
---> problem
1) presence of inhibitors
2) inefficient recovery ; ∥~50% 이상, in less than 5 kb
∥ no satisfactory more than 5 kb (size)
∥ no satisfactory, less than 500ng DNA (amount)
---> methods
1) electrophoresis onto DEAE-cellulose membrane
2) electroelution into dialysis bag
3) low-melting gel
(1) DEAE-cellulose membrane
⇒ simultaneously many samples, constant high yield (0.5-5 kb)
⇒ high purity (microinjection)
⇒ using high ionic strength buffer, eluted
⇒ 15 kb > fragment is not available
(2) dialysis bag
⇒ for the large fragment (5 kb > )
(3) low-melting gel
⇒ less producible, direct ligation after digestion
(C) polyacrylamide gel
---> a monomer
↓free radicals (TEMED, ammonium persulfate)
polymerization
↓methylenebisacrylamide (1/29)
cross-linking, gel
---> advantages
1) resolution is more good
2) accommodate larger quantities of DNA
3) purified DNA is so pure
---> kinds
1) non-denaturing gel ; can't discern the size of dsDNAs (10% difference)
2) denaturing gel ; in presence of urea, formamide
for isolation of ssDNA labeled probe
for analysis of the products of DNA sequencing reactions
3장; 단백질 구조, 기능과 연관된 실험적 분석
(1) How the activites of cellular proteins are regulated ?
1. 이유; 단백질별 수명이 다양함 예) cyclin (분), lens of the eye (나이)
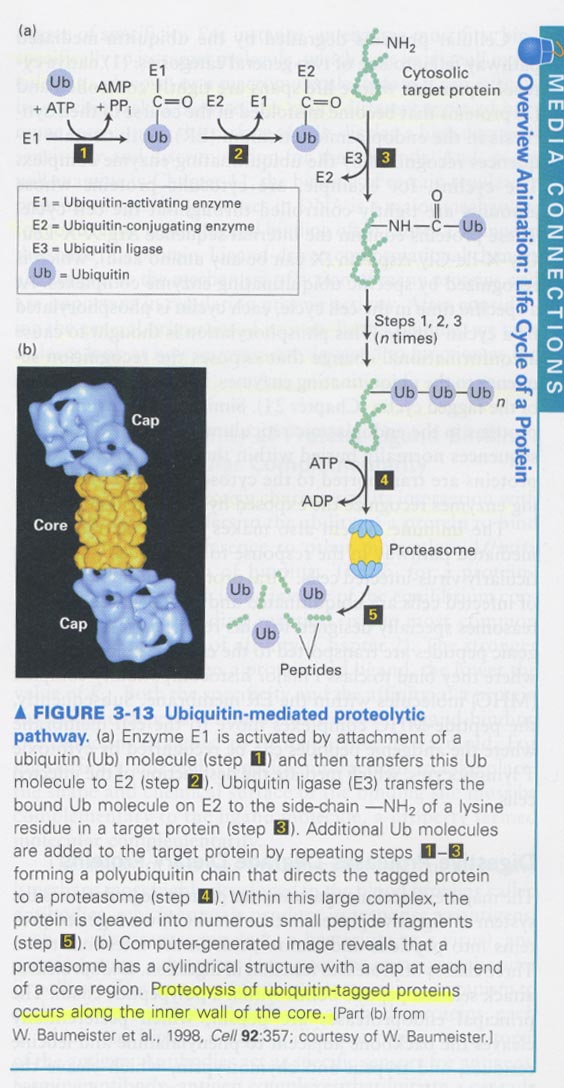
2. Ubiquitination & Lysosomal degradation
a) Lysosomal degradation --> for extracellular proteins and aged or defective organelles
b) ubiquitination --> for proteins whose life spans are tightly controlled
for proteins that becomes misfolded
주요인자: recognition sequence; Arg-X-X-Leu-Gly-X-Ile-Gly-Asp/Asn
phosphorylation (cyclin), exposure of hydrophobic seq (misfolded protein in ER)
예) immune system -- viral proteins of virus-infected cells
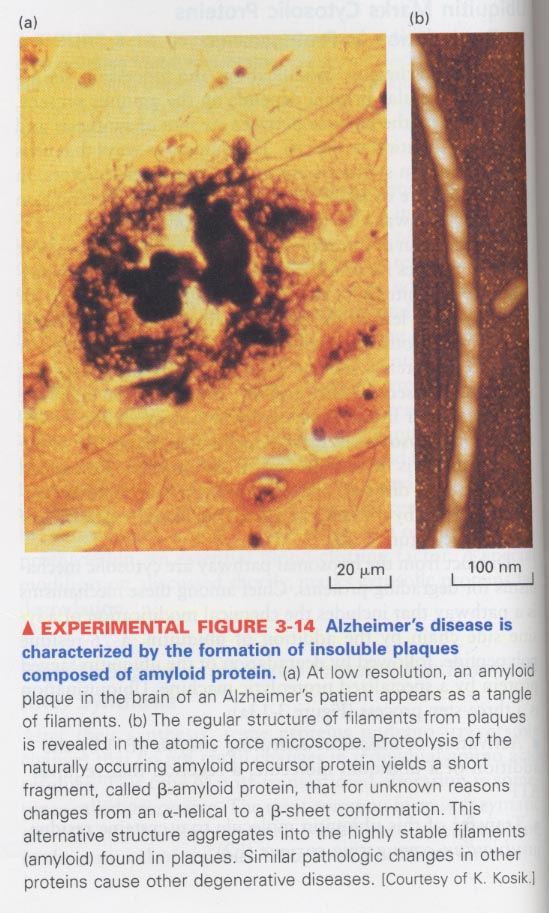
(2) Disease implicated in misfolded proteins
--> proteolytic degradation --> presence of insoluble proteins in various organs
예) Alzheimer's disease, Parkinson's disease, mad cow disease
-- β-amyloid protein 축적
(3) Purification methods of proteins
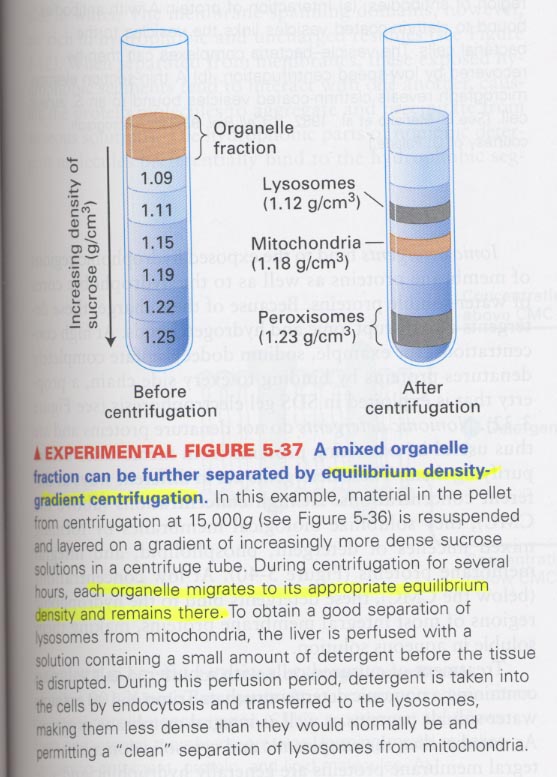
1. centrifugation
원리; size and density
종류; ① differential centrifugation; separation of soluble proteins from insoluble materials
② rate-zonal centrifugation; size(mass) 이용 --> sucrose solution, for separating of many different types of
polymers and particles

③ equilibrium density-gradient centrifugation; for separating of DNA or organelles
2. electrophoresis
원리; charge:mass ratio
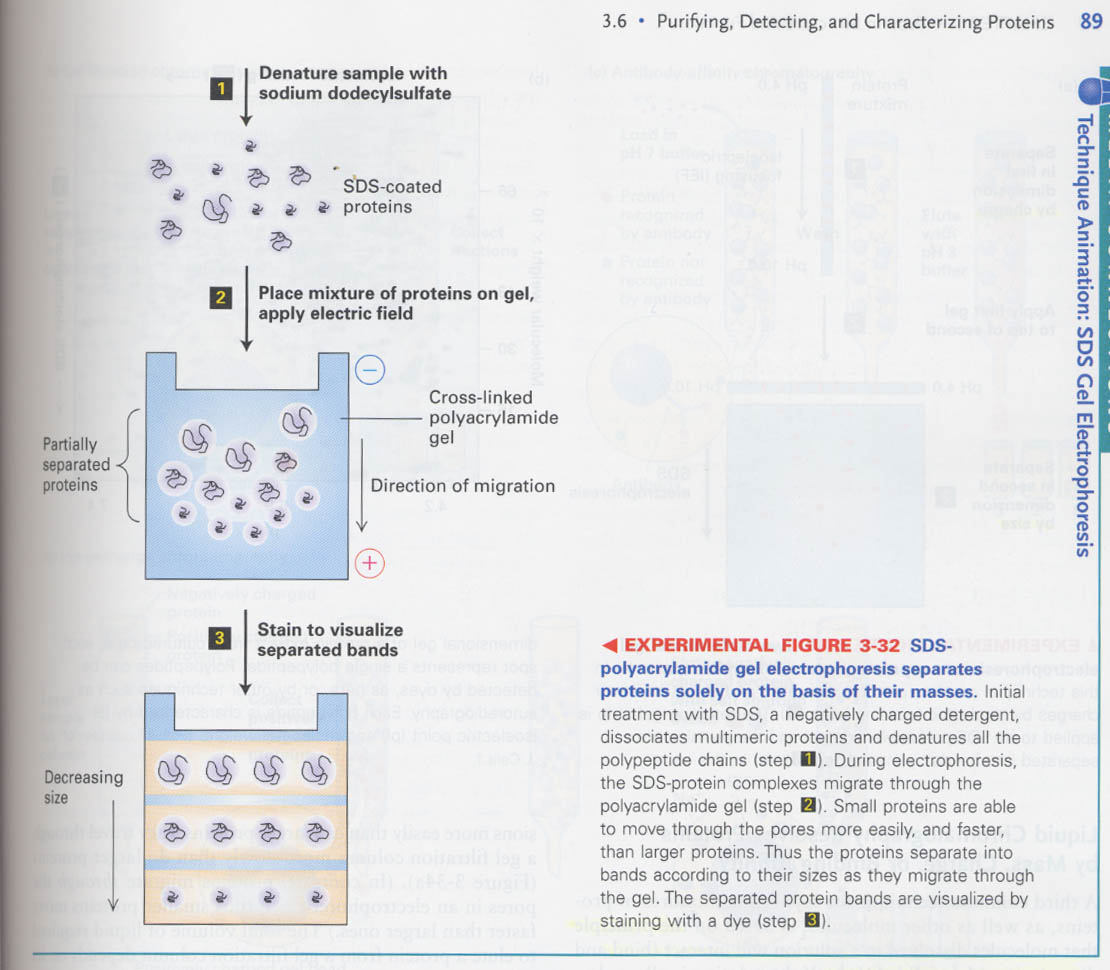
① SDS-PAGE
--> rate is determined by pore sizes and the strength of the electric field
--> SDS효과; chain length, not shape, is a sole determinant
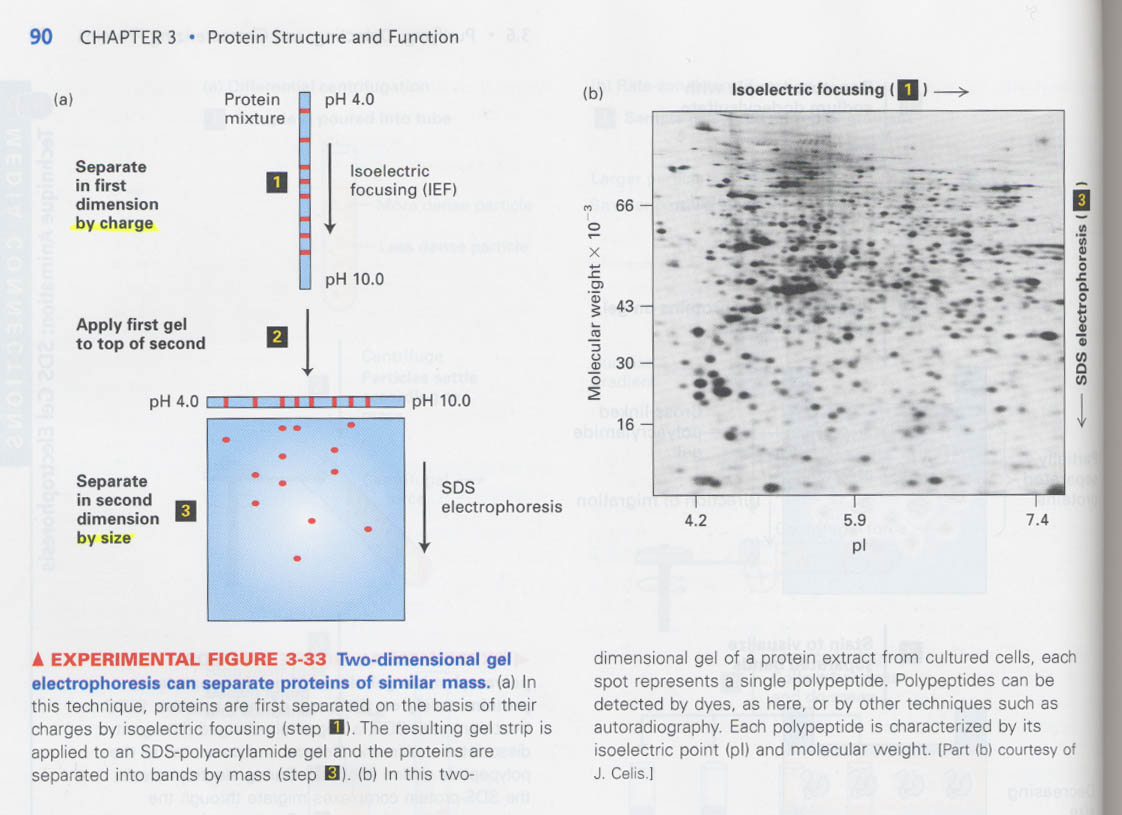
② two-dimensional gel electrophoresis
목적: 아주 유사한 분자량을 가진 단백질의 분리 예) 41KDa & 42KDa
원리: charge and mass
charge에 따른 분리 --> pH gradient, mass에 의한 분리 --> 전기영동
용도: 세포분화/미분화, 암/정상세포간의 단백질 차이분석, 1000 proteins을 동시에 분석
3. chromatography
원리; mass, charge, binding affinity에 의한 분리
① gel filtration
beads --> polyacrylamide, dextran, agarose
무거운 것일수록 빨리 빠져나옴
② ion-exchange chromatography
beads --> positive charge or negative charge
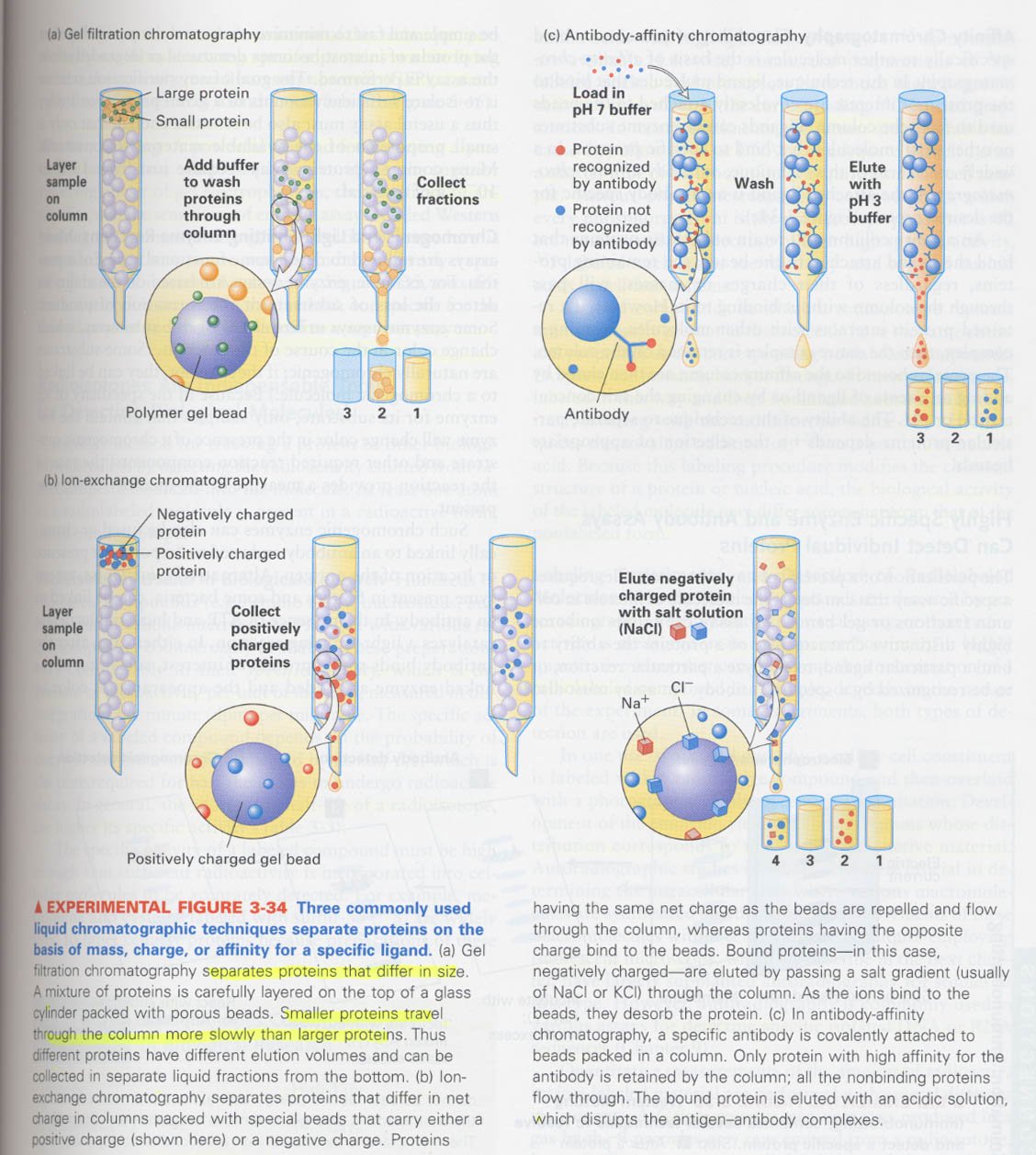
③ affinity chromatography
beads --> covalently attached with ligand
elution --> by adding of excess of ligand or changing of salt concentration or pH
(4) identification of the interested proteins
Assays; simple, fast, minimal error, no degradation of proteins of interest, small amounts of materials (sensitivity)
① chromogenic and light-emitting enzyme reactions
chromogenic --> 반응중 기질의 색깔변화 추적
light-emitting --> luciferase linked to antibody
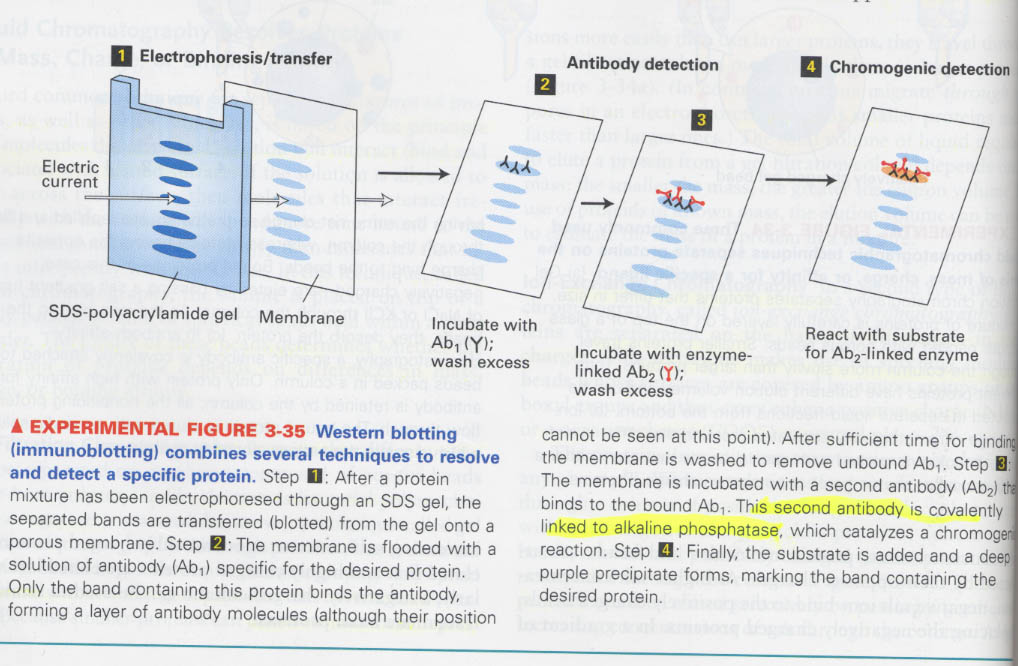
② western blotting
원리; 전기영동의 분리능력 + 항체 특이성 + enzyme assay의 sensitivity
(5) 방사선 동위원소의 실험적 이용
specific activity = the amount of radioactivity per unit of material
shorter the half-life is higher specific activity --> 이유; shorter time of incorporation, smaller cell sample
in general, biological activity between labeled and unlabeled molecules is identical except for 125I-labeled molecules
detection; autoradiography, counter (Geiger counter, scintillation counter), phosphoimager
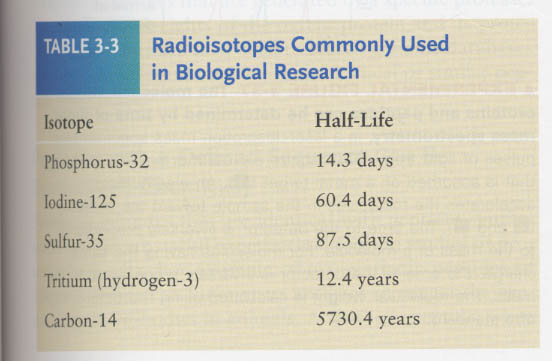
Pulse-Chase experiment; for tracing the location of intracellular proteins
for tracing the transformation of metabolite into others over time
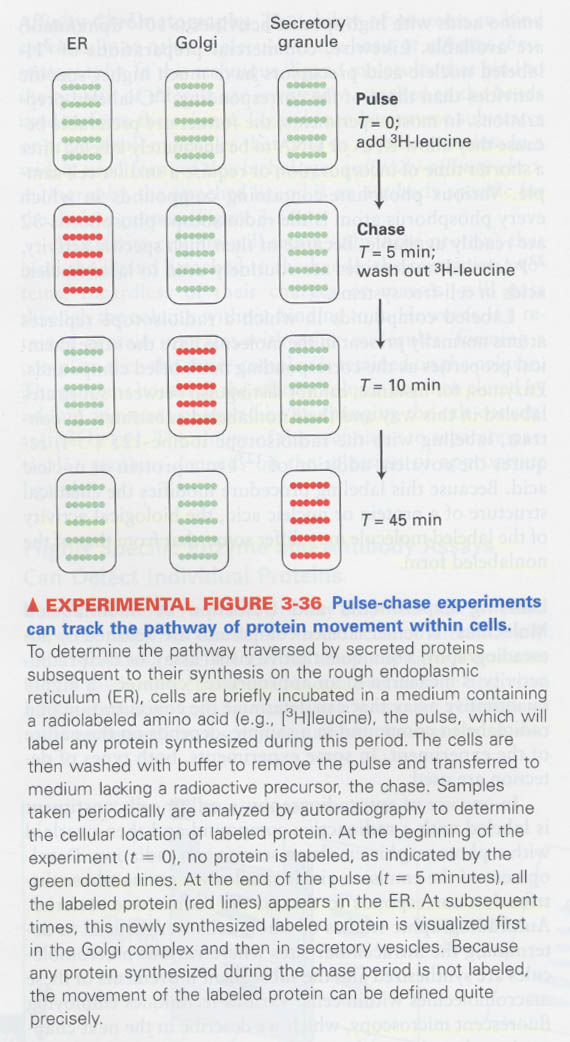
(6) Mass spectrometry; for measuring the mass of proteins
1 X 10-15 mole, 200,000 MW, 0.1% error
4장; Basic molecular genetic mechanisms
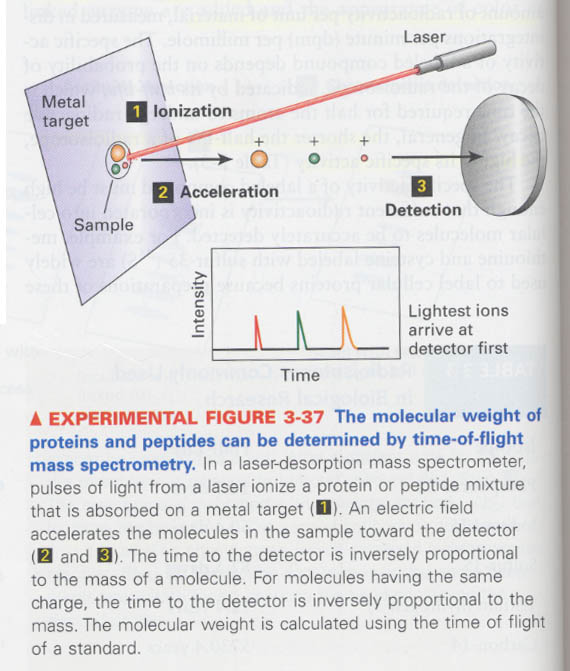
(1) How we can differentiate ds-DNA from ss-DNA ?
Tm ?
denaturation; breakage of H-bonding and others, when; replication and transcription
(2) How we can measure the G:C content of DNA ?
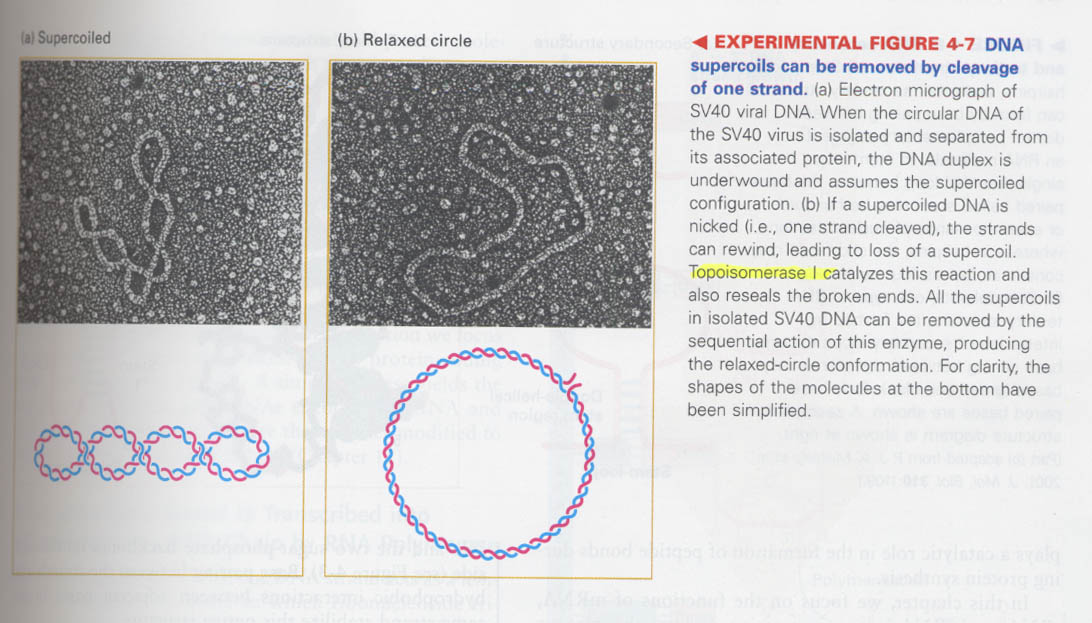
(3) How do you know that DNA is circular and supercoiled ?
topoisomerase I --> induce and release supercoiling
topoisomerase II --> ?
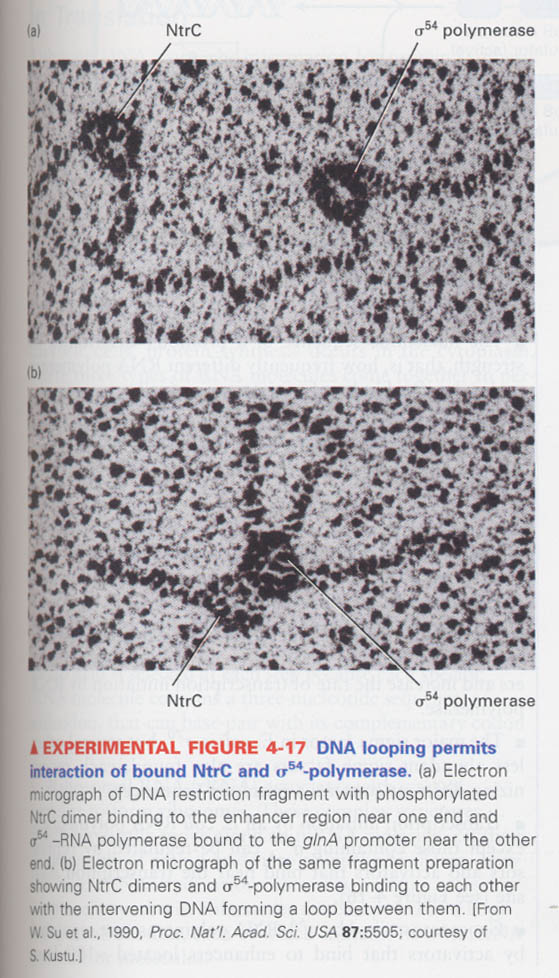
(4) How do we know that activator bound to enhancer interacts with RNA polymerase to initiate the transcription ?
예) σ54-RNA polymerase and its activator NtrC (nitrogen regulatory protein C)
NtrB --> NtrC phosphorylation --> binding to enhancer of GlnA gene --> interacts with σ54-RNA polymerase
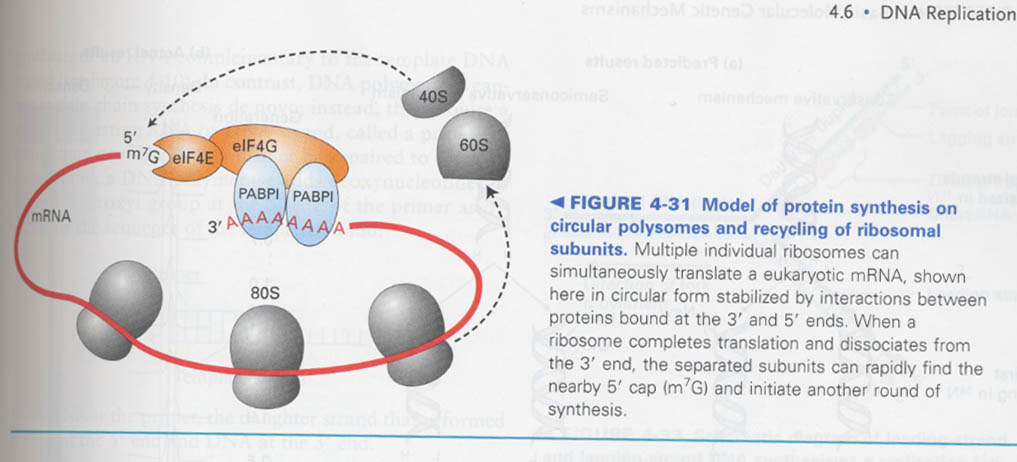
(5) How an eukaryote increases the efficiency of translation ?
--> by polysomes and rapid ribosome recycling (= circular structure of mRNA)
ex) PABP1(polyA-binding protein) + eIF4G
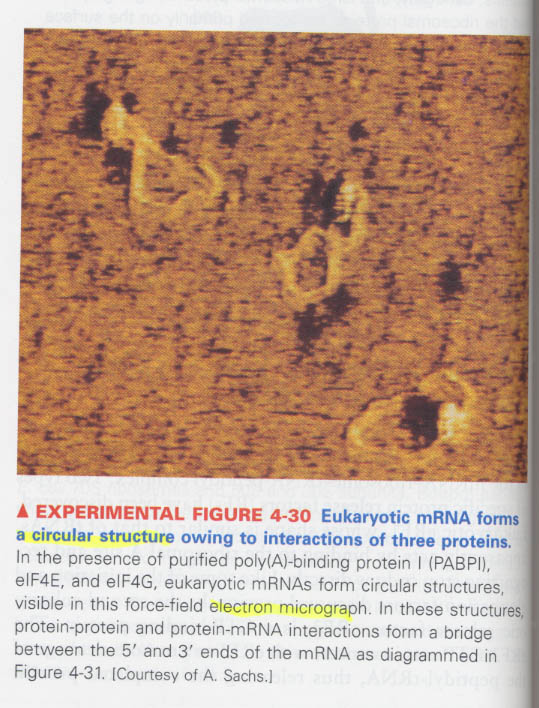
-- Model for a circular structure & polysome
(6) How do we know that DNA replication has a semiconservative replication mode ?
--> growing in the presence of 15N and 14N
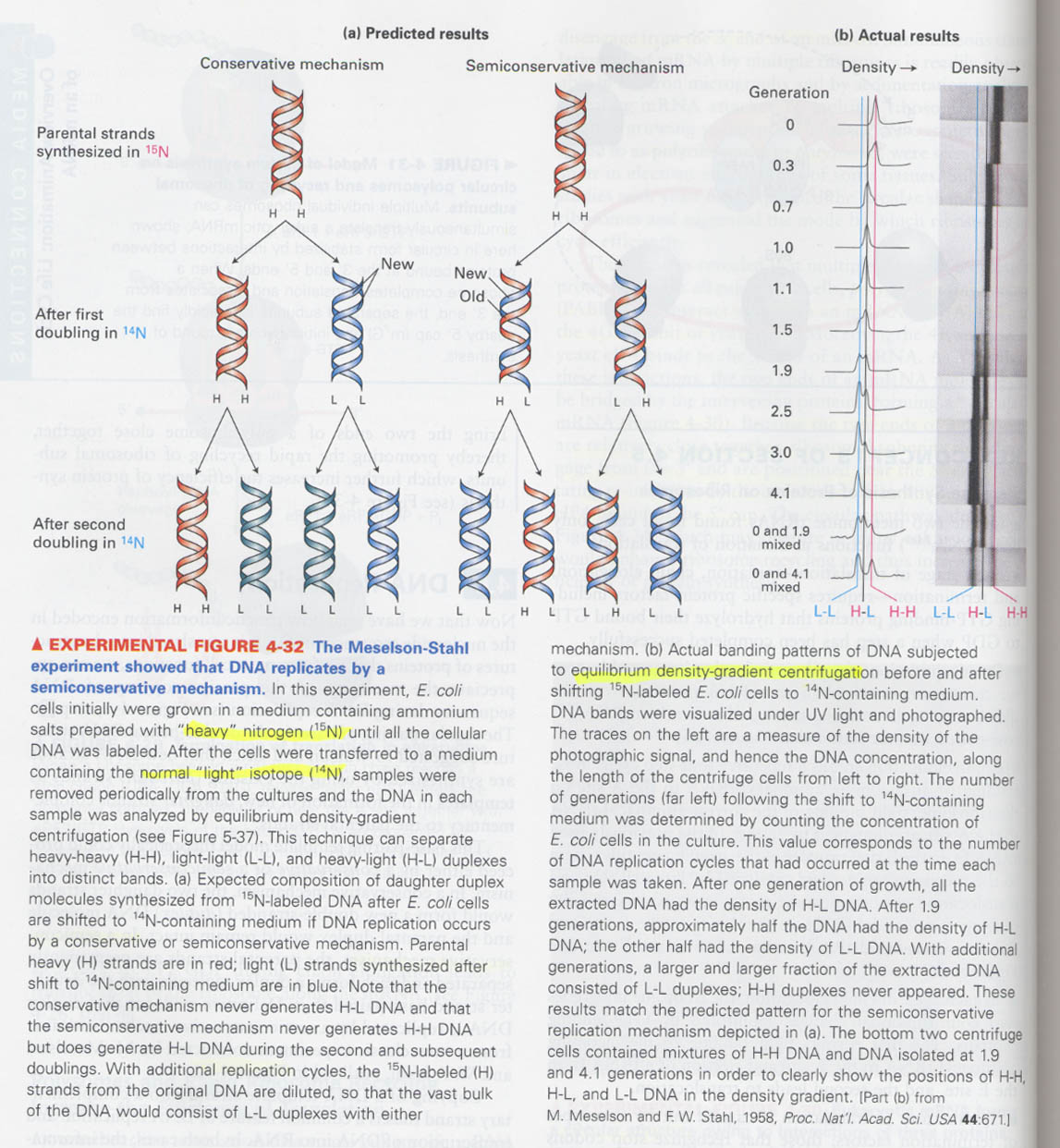
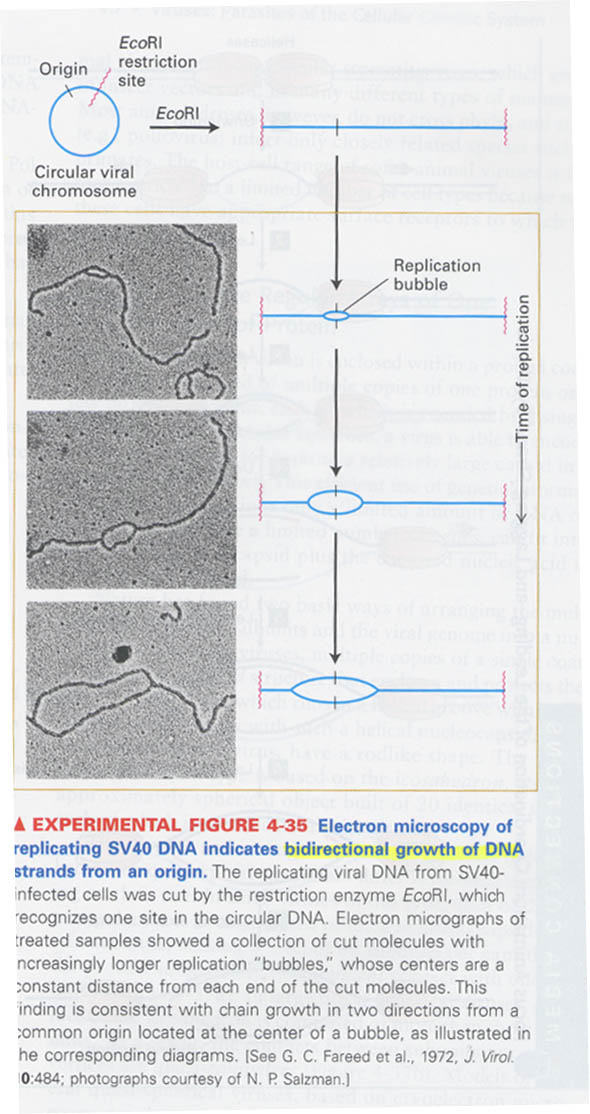
(7) How we can determine that DNA is bidirectionally replicated ?
5장; Biomembranes & Cell architecture
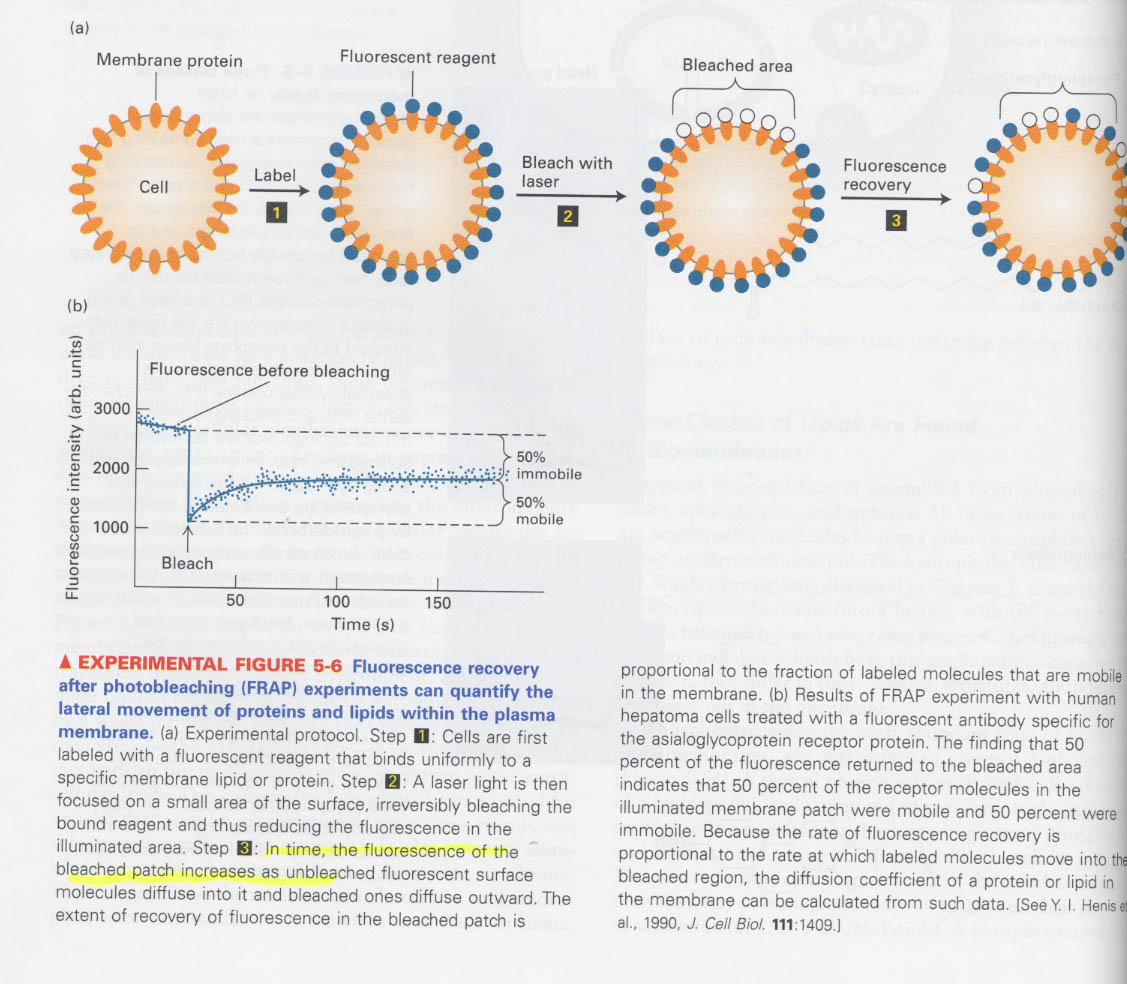
(1) membrane상의 lipid와 단백질은 lateral diffusion을 행한다.
--> 107 times/sec, several μm/sec
--> fluoresence recovery after photobleaching (FRAP) technique
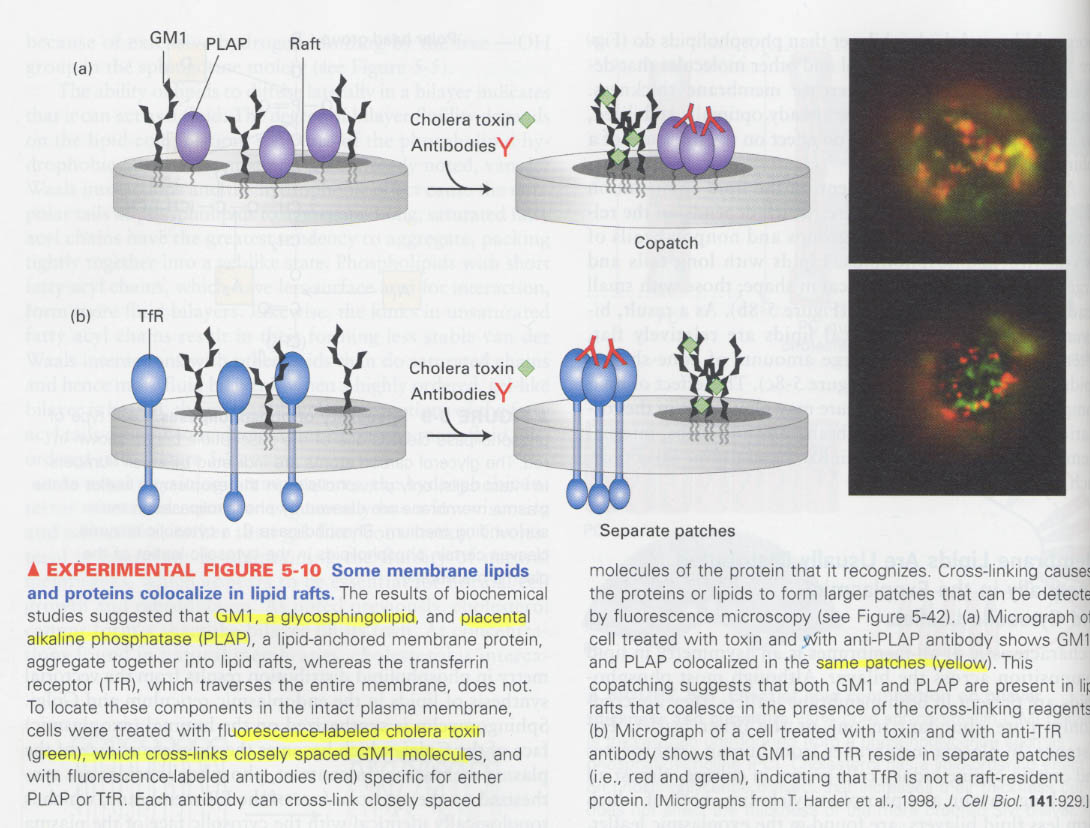
(2) lipid rafts (=microdomain)
--> membrane lipid (cholesterol, sphingolipid)와 protein은 특정부위에 움직이지 않고 한정되어 있을 수 있음
--> How the rafts are destroyed? ① methyl-β-cyclodextrin; depletion of cholesterol
② filipin; sequestering of cholesterol
(3) How different cell types are purified ?
--> fluorescence activated cell sorter ( FACS); by measuring the emitted fluorescent light and scattered light
negative charges propotional to the amount of fluorescence
그밖의 용도; ① DNA/RNA 양측정 ② shape/size 측정
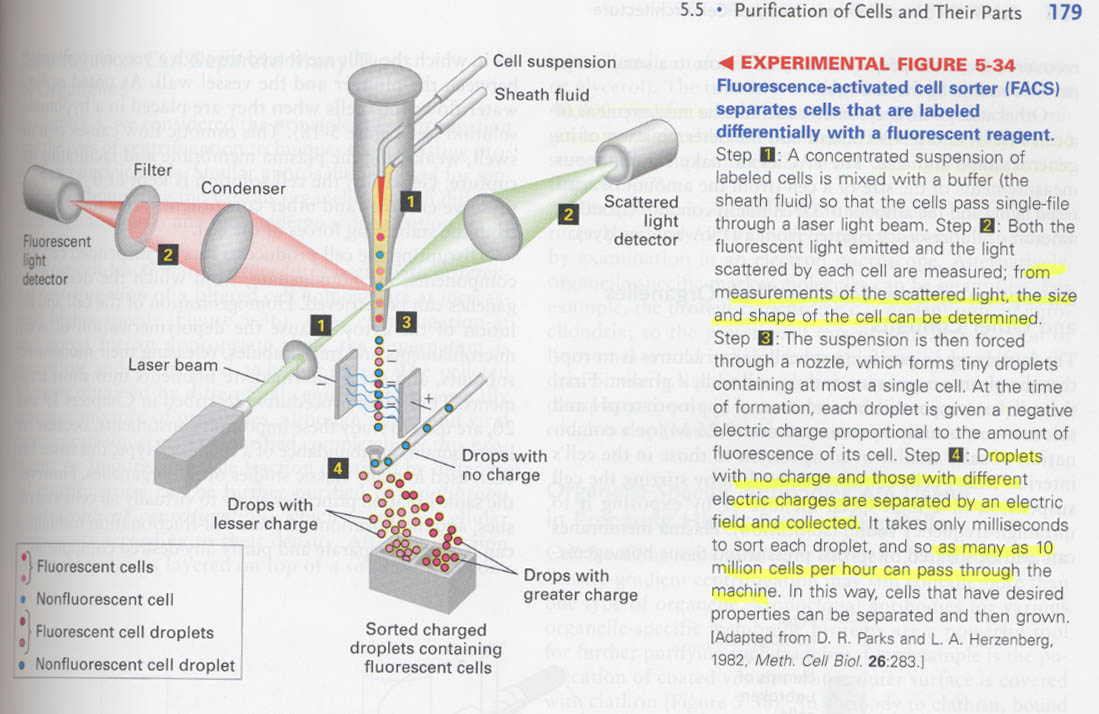
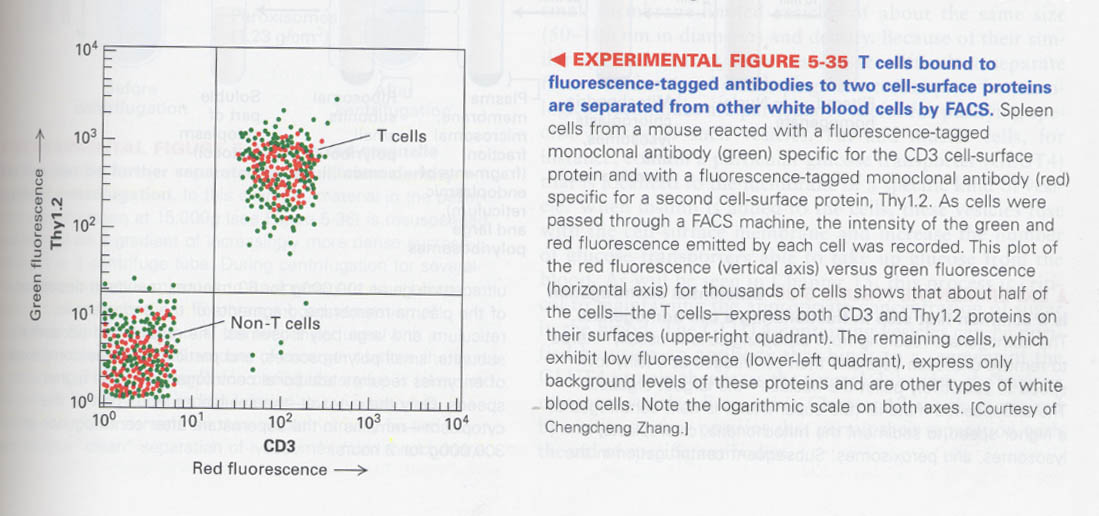
--> 실제 실험결과 (green;anti-CD3, red;Thy1.2.)
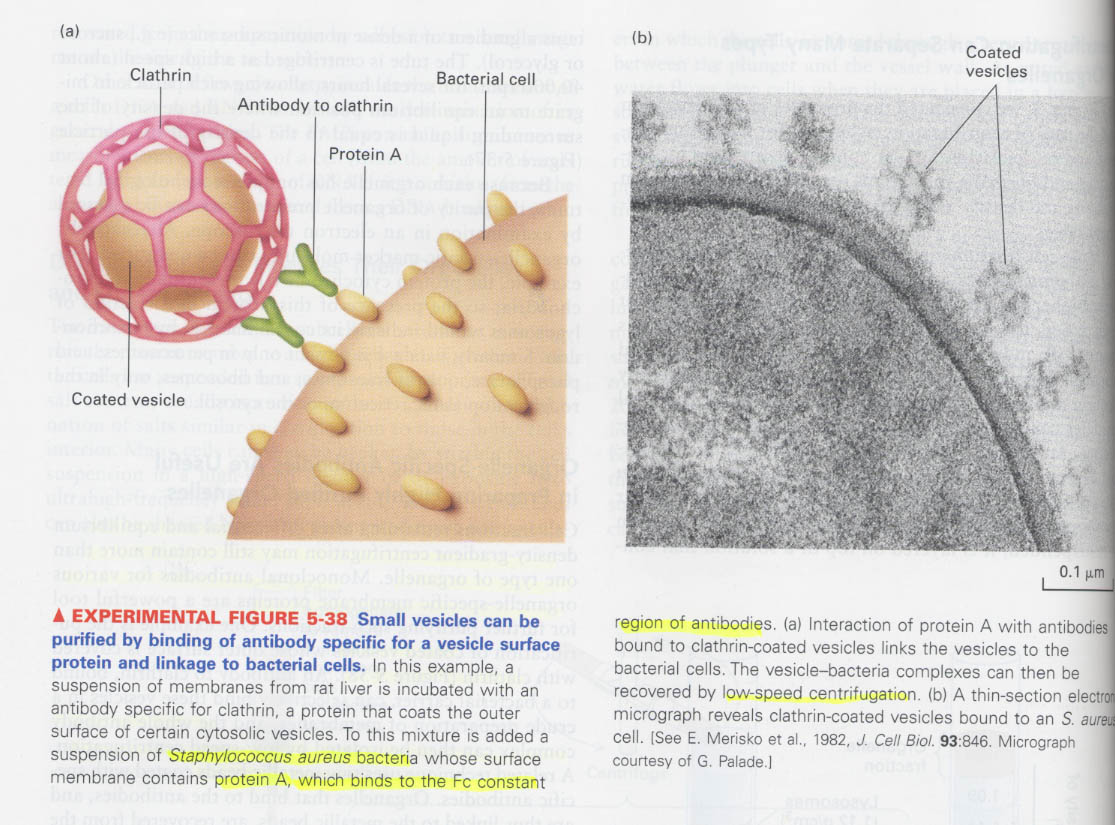
(4) How the specific organelle is purified ?
(예) membraneous organelles; coated vesicles, GLUT4-containing vesicles
--> organelle-specific membrane protein에 대한 항체이용
--> by low-speed centrifugation or metallic beads coated with antibody
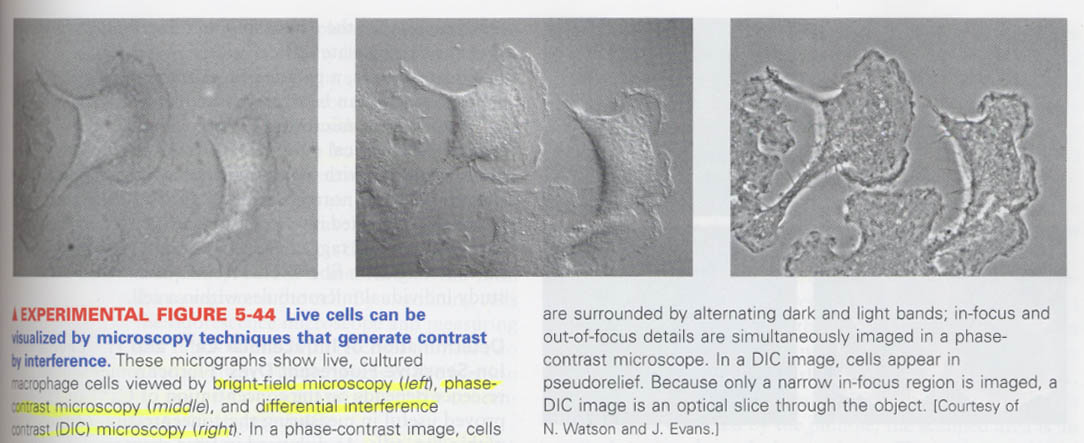
(5) Visualization of cell architecture (by microscope)
--> how the live cells can be measured ? without staining
원리; refractive index, thickness
① phase contrast microscopy ② differential interference contrast microscopy
③ fluorescence microscopy (in fixed cells)
--> for localization of proteins within a cell
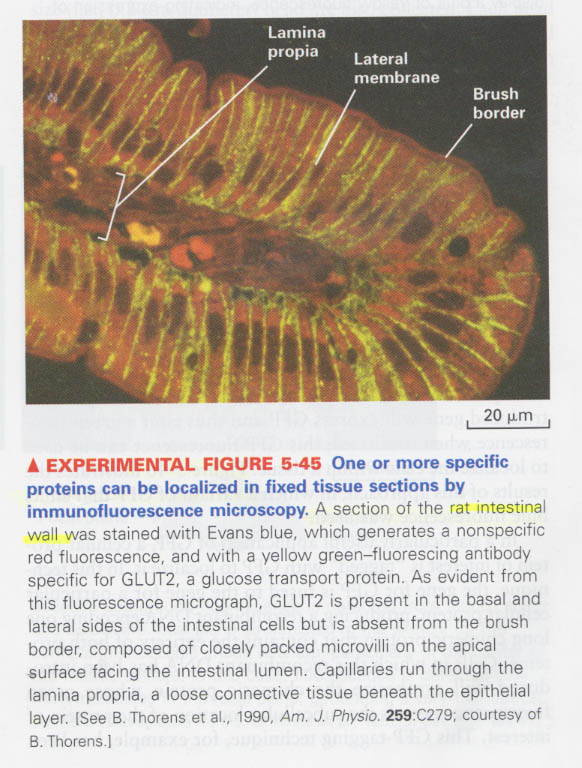
--> flurochrome; rhodamine and Texas red (Red), Cy3 (orange), fluorescein (green)
--> 예) GLUT2
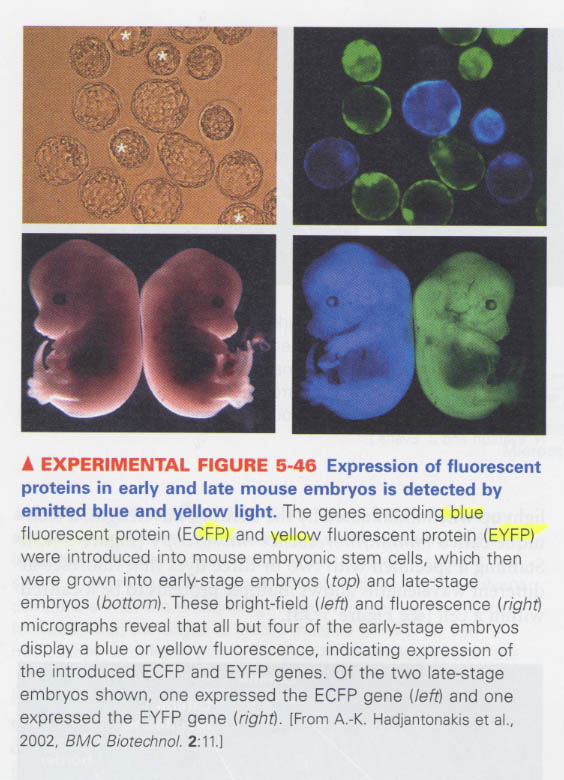
④ fluorescence microscopy (in live cells)
--> introduction of genes such as GFP (green), CFP (blue), YFP (yellow)
--> 예) mouse embryo
 **
Yellow --> green
**
Yellow --> green
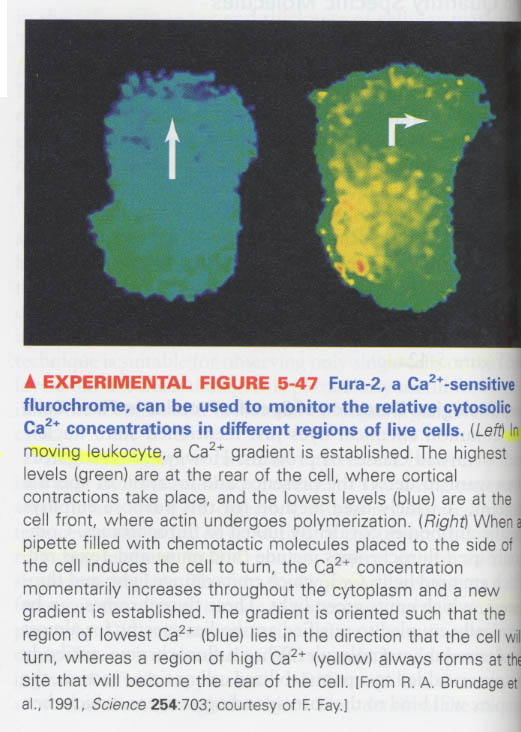
--> 예) 세포내 Ca2+와 H+양의 측정; ion-sensitive fluoresent dye 이용
ⓐ fura-2 ; Ca2+ sensitive dye
fura-2 + ethanol -> fura-2 ester -> lipophilic -> in cytosol, hydrolysis of ester -> no transfer
ⓑ SNARF-1 ; H+ sensitive dye
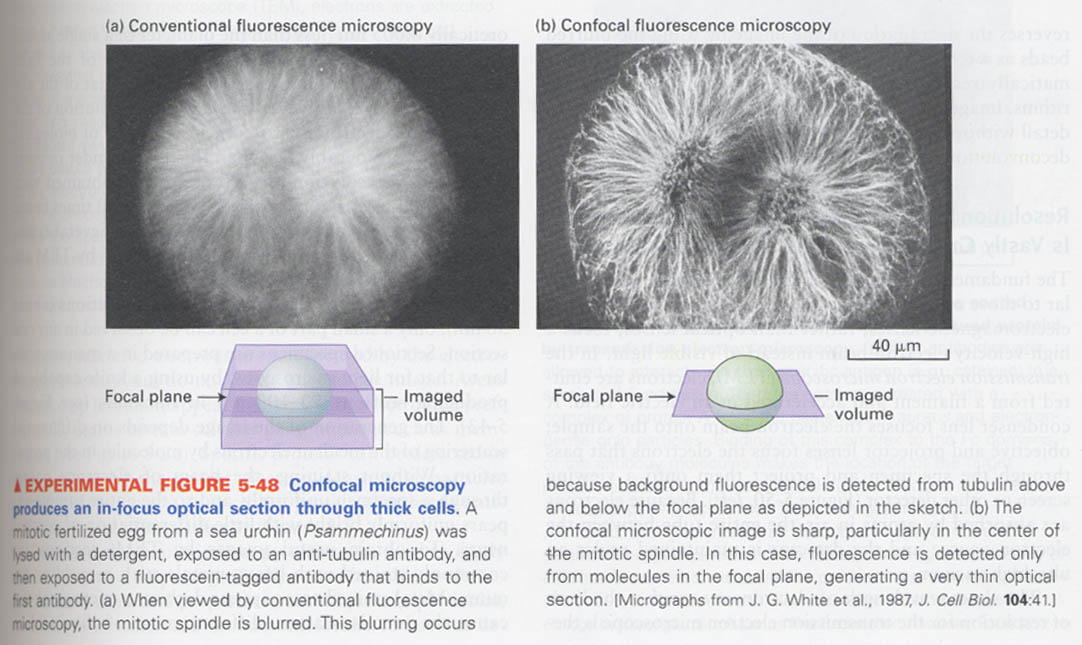
⑤ confocal scanning
limitations of conventional fluorescence microscopy --> 1) destroy materials during processing of cutting a section
2) fluorescence light emitted from molecules above and below
the plane of focus
⑥ deconvolution microscopy
--> same image-sharpening effect, but through a different process
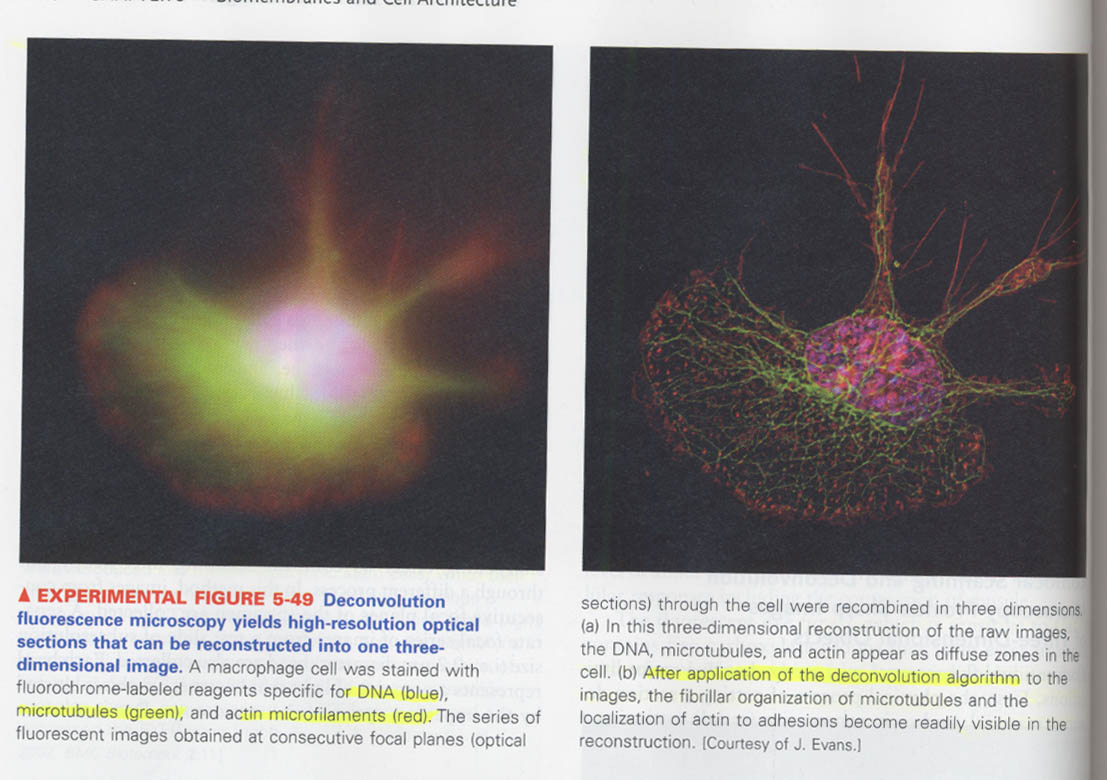
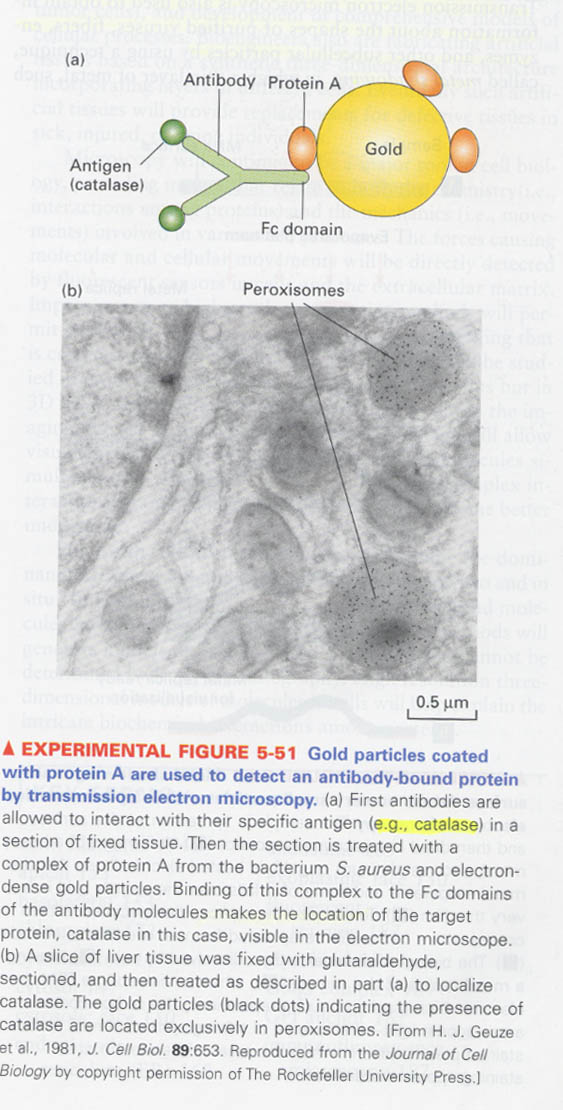
⑦ transmission electron microscopy (TEM)
--> very thin and fixed section (50nm), only a small part of a cell
--> detection of specific proteins in the thin sections ; use of electron-dense gold particles
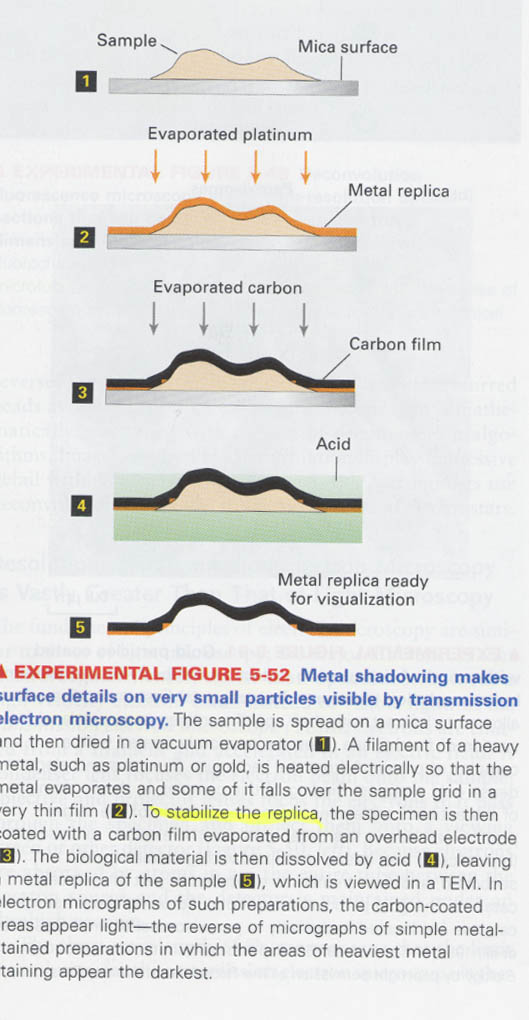
metal shadowing; for detection the shape and its component of a cell
steps; 2) making a metal film, 3) stabilization of replica
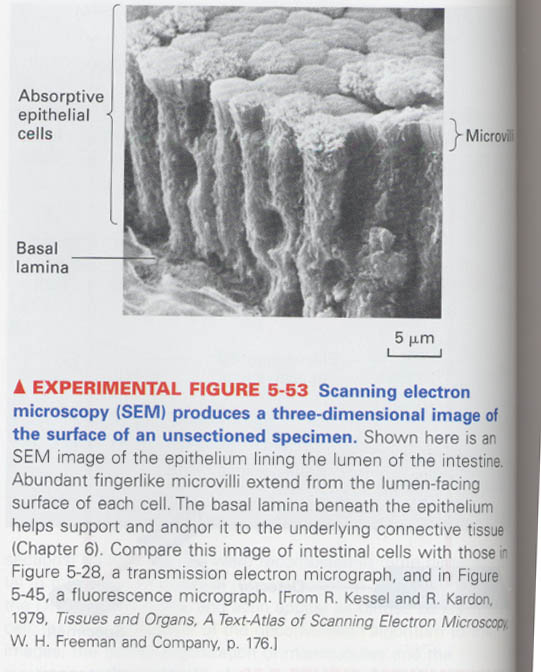
⑧ scanning electron microscopy (SEM)
--> only 10nm thickness, surface of unsectioned metal-coated specimens
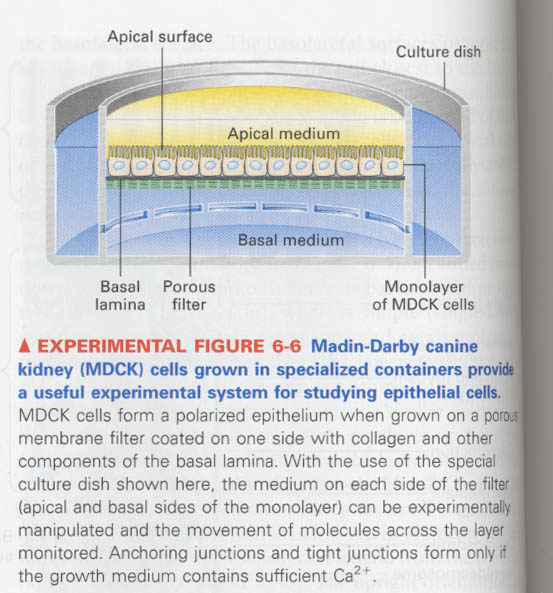
6장; Cells into Tissues
(1) Cadherins
--> 6 subfamilies, 100 이상 proteins, classical cadherins (E-, P-, N-)
--> cell-cell adhesion, signaling, tissue differentiation
--> Ca2+ dependent adhesion
--> E-cadherin의 역할을 규명하기 위한 실험방법; media change, antibody 이용
① Structure of cadherins
cadherin domain; for Ca2+ binding (-> rigidify the cadherin oligomers) and cell-cell adhesion (3 cadherin domains 관여)
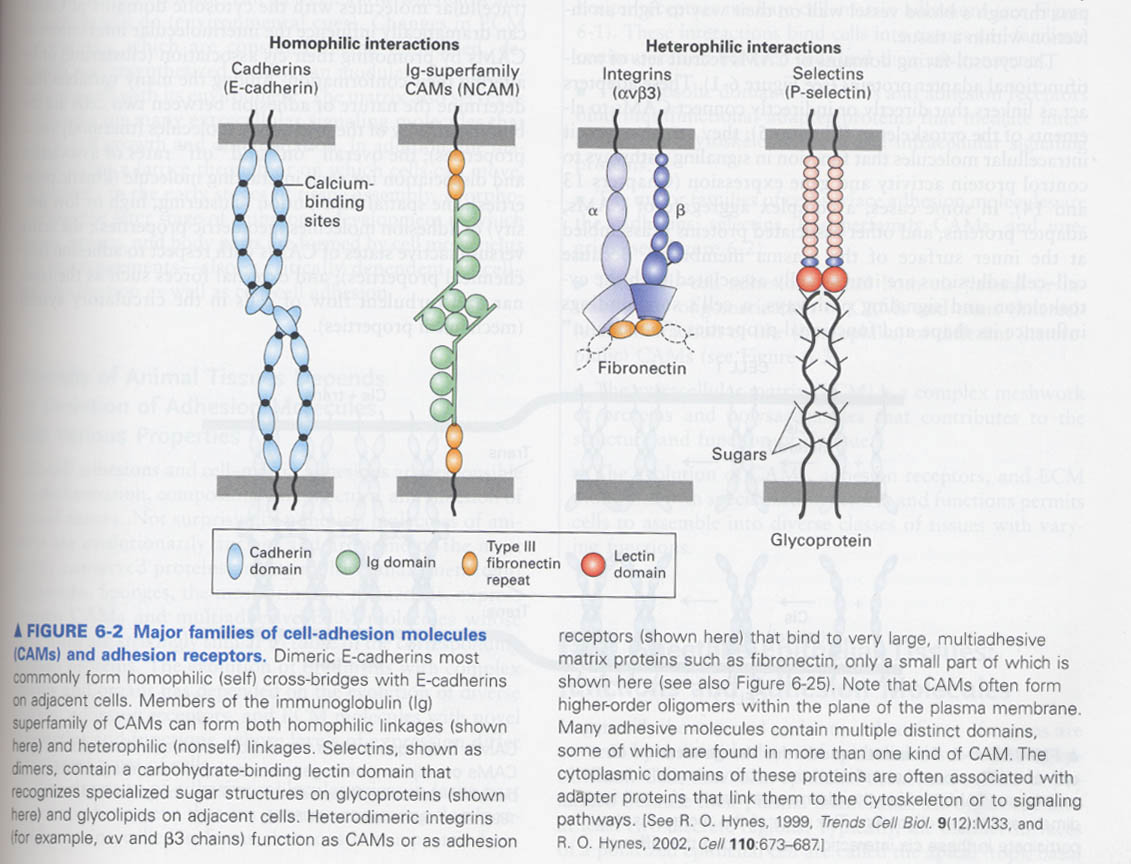
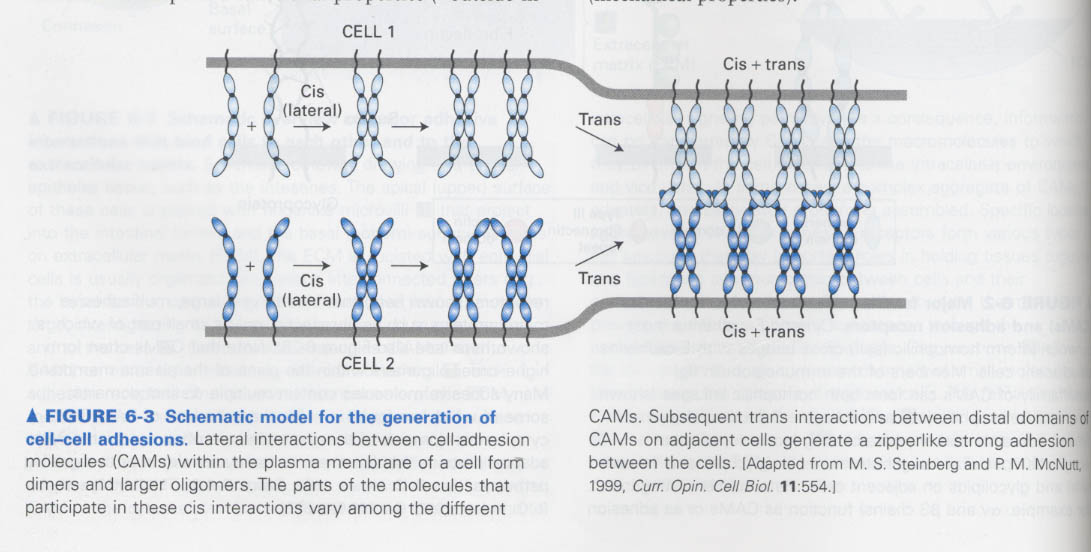
How cadherins mediate the cell-cell interactions ?
--> by cis or trans interactions
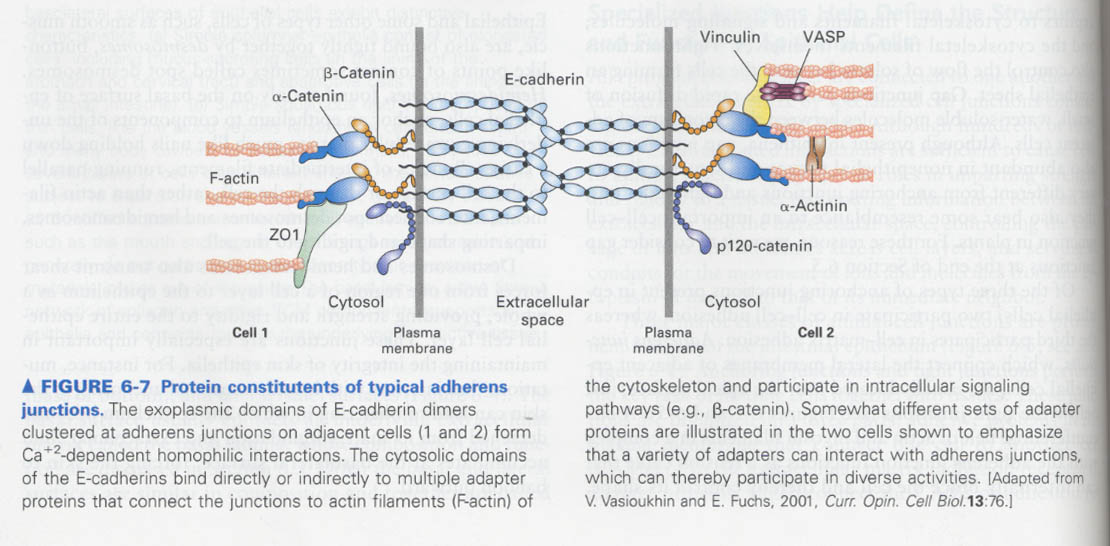
Where the C-terminal of cadherins is linked with ?
--> actin cytoskeleton and cytosolic adaptor proteins
--> in tumors; defect in this interaction
② tight junction
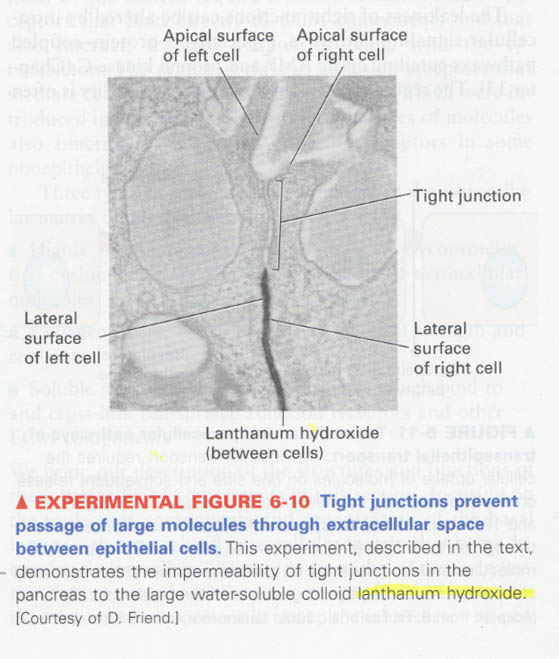
--> impermeability of water-soluble substances
structure; Fig 6-9, JAM (junction adhesion molecule)
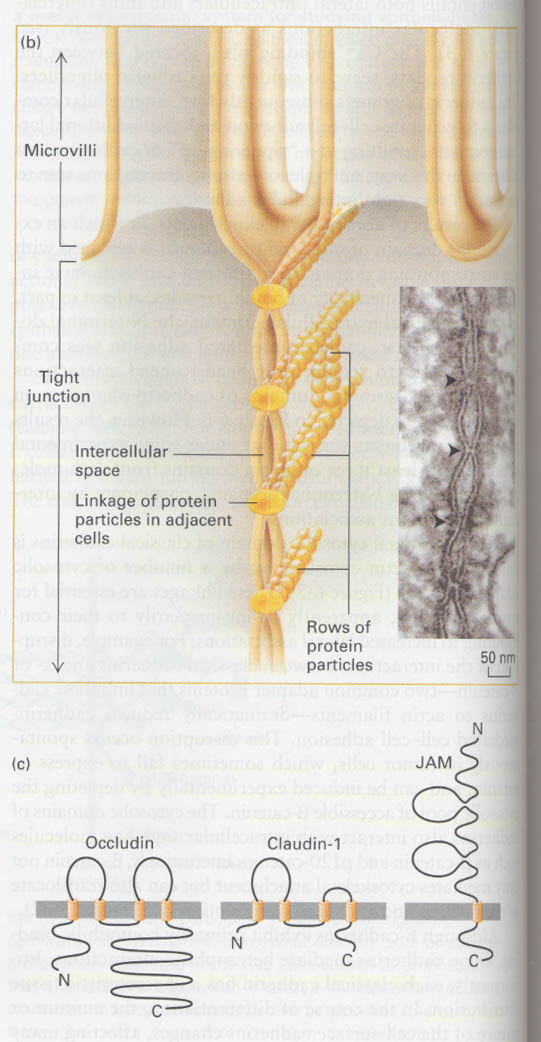
Experiment for demonstration of impermeability of water-soluble substances in tight junction
--> use of lanthanum hydroxide (electron-dense colloid)
--> importance of Ca2+ in the formation and integrity of tight junctions
if a low conc. of Ca2+ in a chamber -- freely movement of fluids and salts
if a high conc. of Ca2+ in a chamber -- no freely movement of fluids and salts

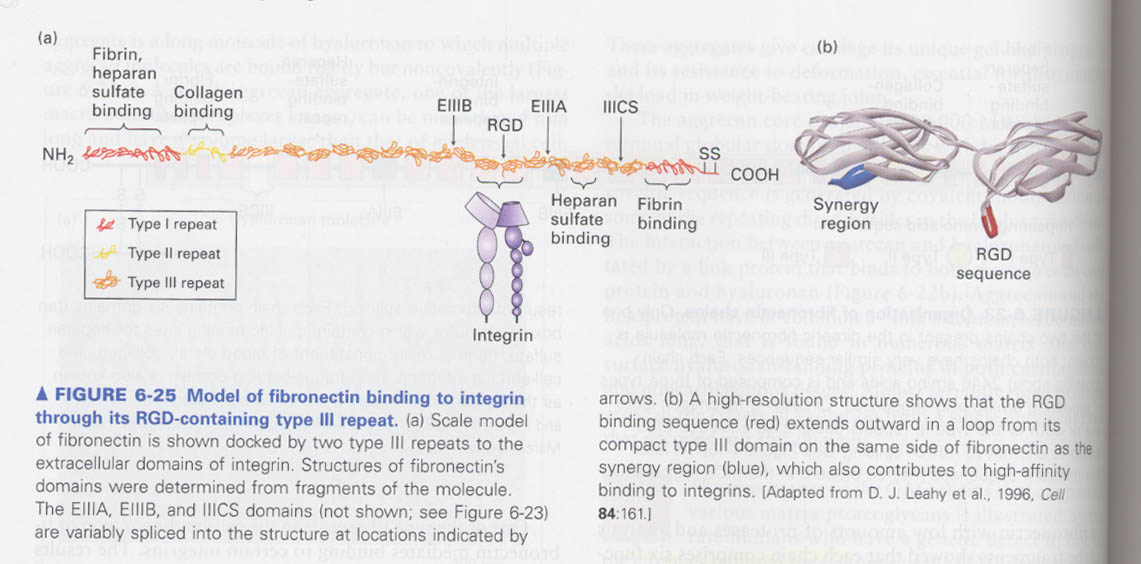
③ fibronectin
--> a matrix protein, cell migration and differentiation, wound healing, a dimer protein (C-terminal linked)
6 functional regions, 3 types repeats
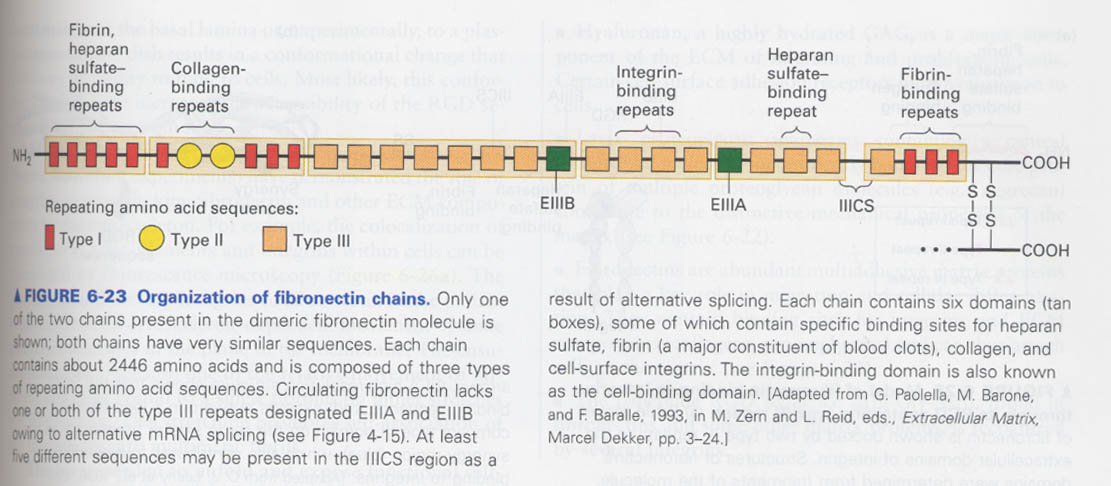
--> intergrin binding sequence of a fibronectin repeat type III; Arg-Gly-Asp (RGD) sequence
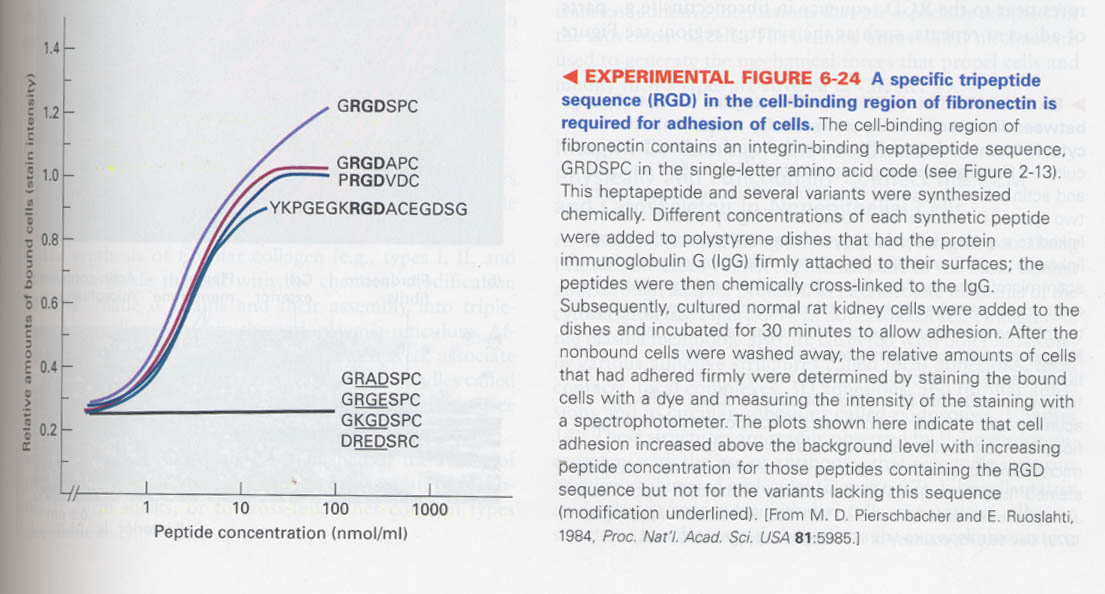
--> Model for fibronectin binding to integrin
synergy region and other RGD-containing proteins enhance the binding
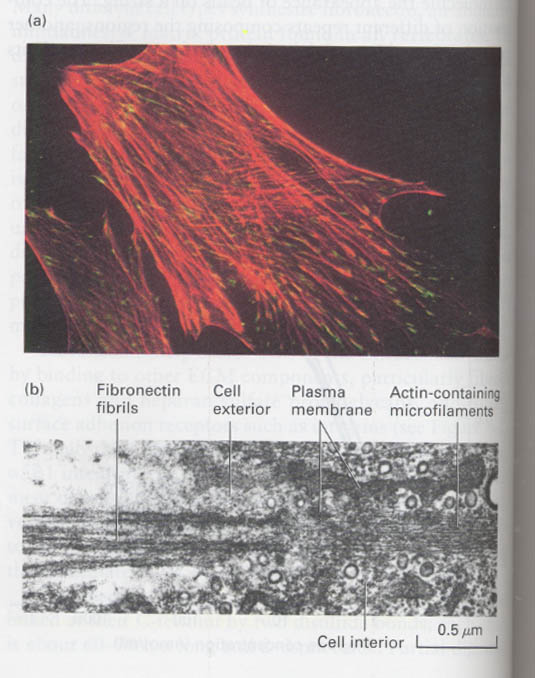
--> colocolization of integrins and actin filaments; red (anti-actin), green (anti-integrin)
④ Hybridoma technique; for prepartion of monoclonal antibodies
How do we make a hybridoma ?
--> fusion; viral glycoproteins, PEG
--> selection media; HAT medium
Use of monoclonal antibodies; affinity chromatography, immunofluorescence microscopy, therapeutic tools
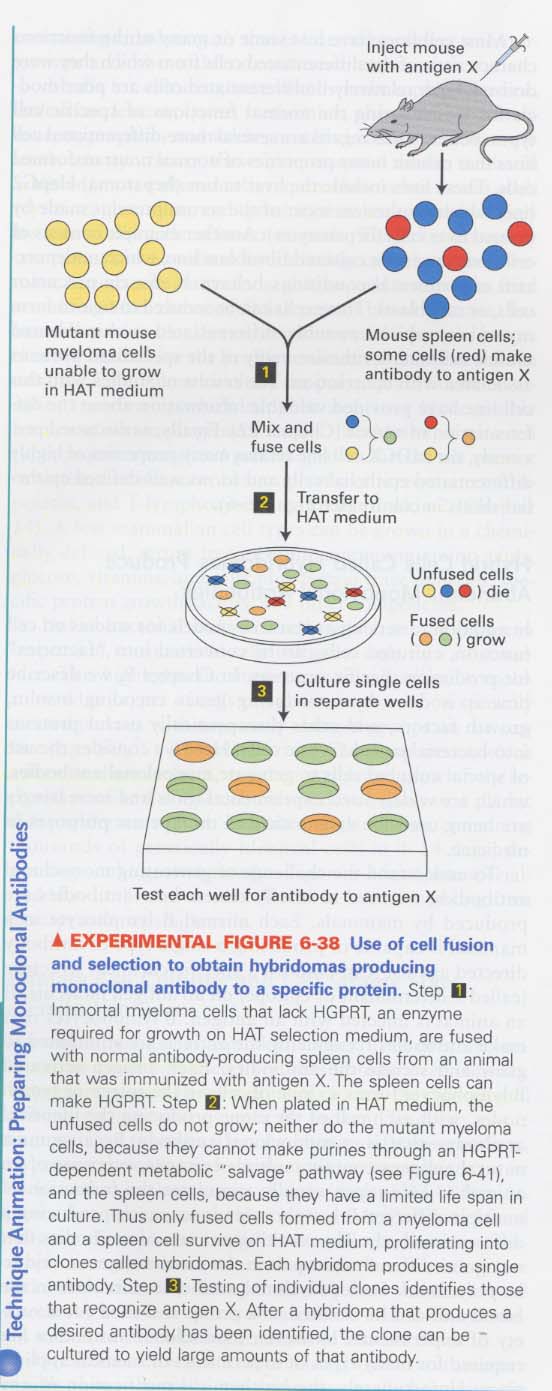
--> HAT medium containing hypoxanthine, aminopterin, thymidine
myeloma cells (HGPRT-) --> die

7장; Transport across Membranes
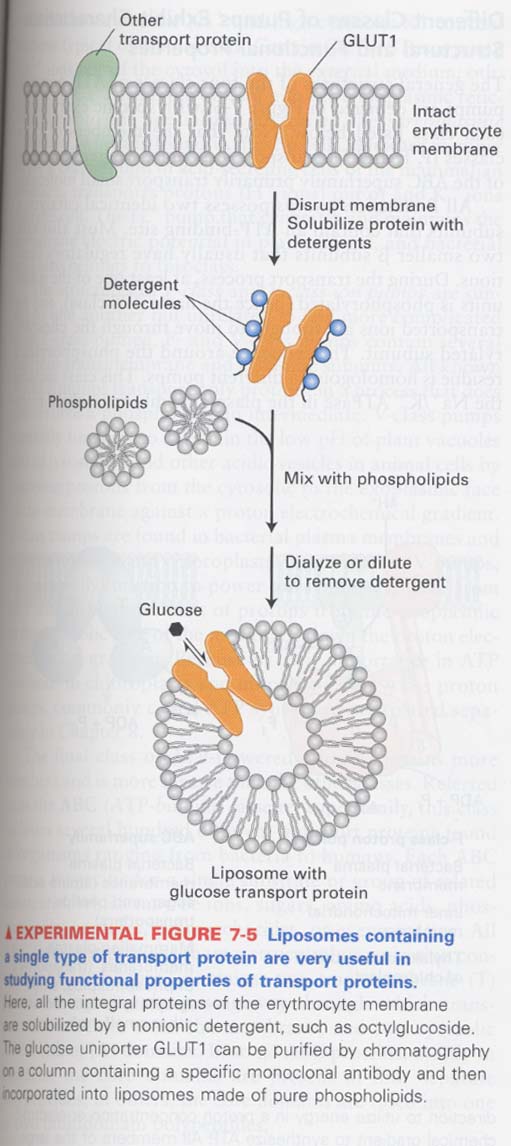
(1) How we study the function of transport proteins ?
① liposome 이용
② transfection into cells that normally do not express transport proteins
(2) How the transmembrane electric potential can be arised ?
--> use of potentiometer

(3) How is membrane electric potential in animal cells ?
--> many K+ channels (outward), a few Na+, Cl-, Ca2+ channels
--> negative charges on the inside, positive charges on the outside
K+ channels: nongated (not affected by membrane potentials or small signaling molecules
--> 실제 membrane potential = -70 mV (by presence of Na+channels (inward))
--> Na+/K+ ATPase
** in plant and fungal cells
--> by by transport of H+ outward
How the membrane potentials are measured ?
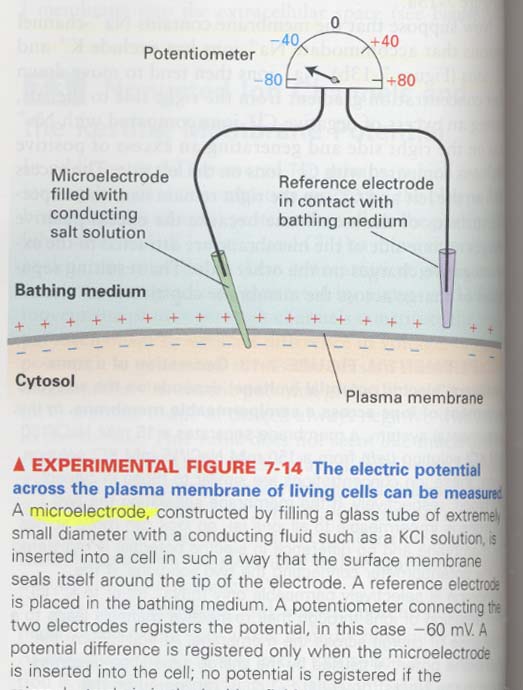
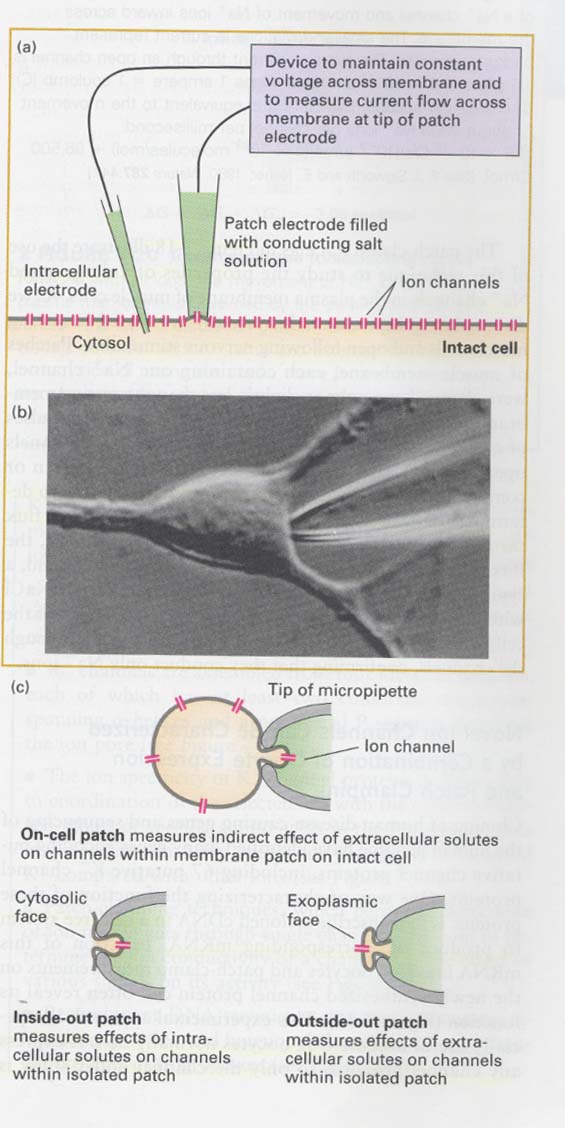
(4) Patch clamping technique
--> to measure ion movements through single channels
--> in whole cells or isolated membrane patches
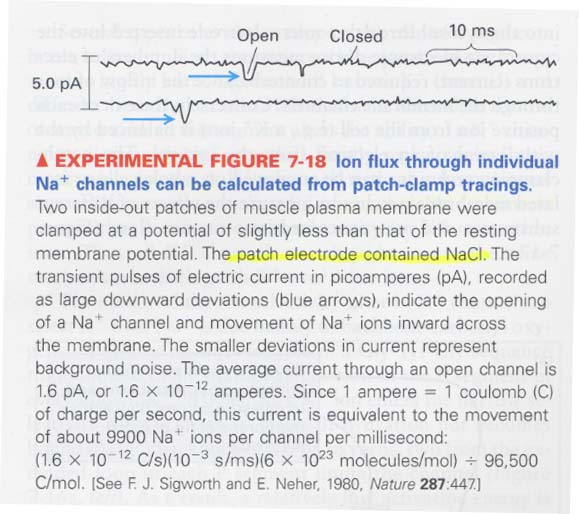
** patch-clamping tracing; measure the time for opening or closing of channels
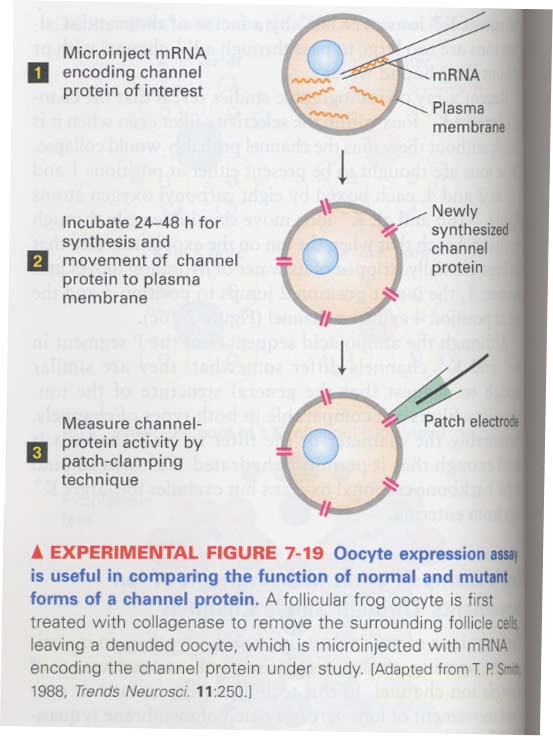
** How novel ion channel proteins are characterized ?
--> in vitro transcription --> oocyte expression --> patch-clamping
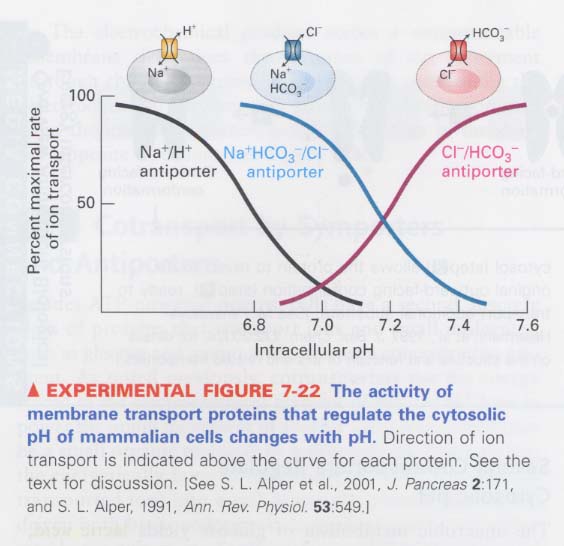
(5) How the cytosolic pH is regulated ?l
--> in metabolism; produce excess H+ ions from H2CO3
--> How the excess H+ ions are removed from cytosol ?
① Na+HCO3-/Cl- antiporter; HCO3- (inward) -> CO2 + OH- (by carbonic anhydrase)
OH- + H+ = H2O
② Na+/H+ antiporter
** How cells cope the excess of OH- under a certain circumstance ?
--> HCO3-/Cl- antiporter 사용하여 HCO3-를 세포밖으로 방출
(5) water movement
aquaporins; water channel proteins, a tetramer
each subunit = 6 membrane-spanning α-helix
How the waters can be passed through the channels ? --> constriction by conserved hydrophilic Aas of side chain
and carbonyl groups
--> by H-bonding between water and amino groups of side chain
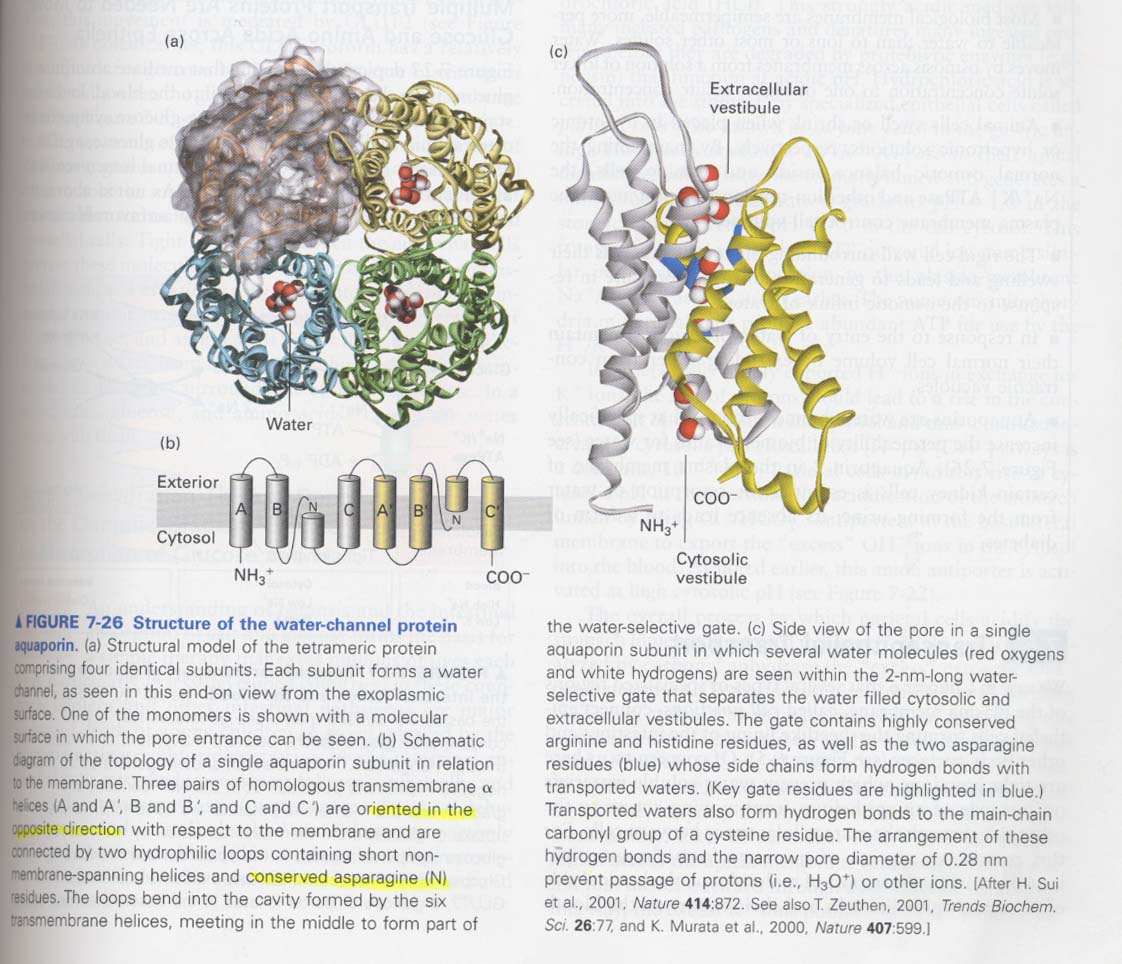
** evidence of aquaporins as water channels

(6) Action potentials in nerve cells
morphology of neurons; ① cell body ② dendrite ③ axon ④ axon terminal

① cell body
--> nucleus, synthesis of all neuronal proteins and membranes
② axon
--> conduction of action potentials, -60 mV (in resting), +50 mV (in stimulus)
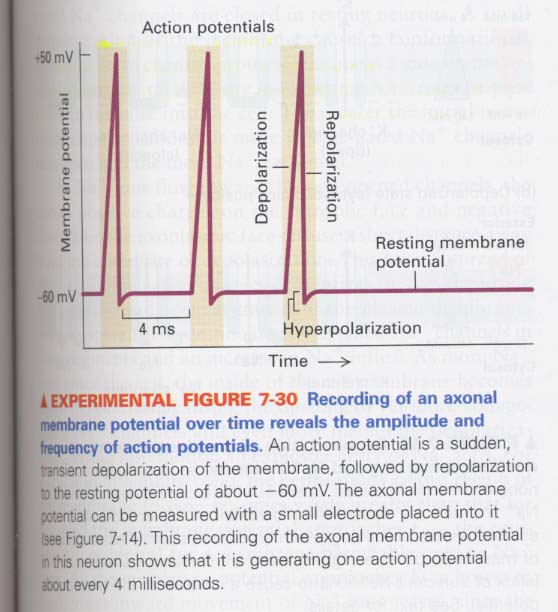
③ axon hillock
--> origin of action potentials
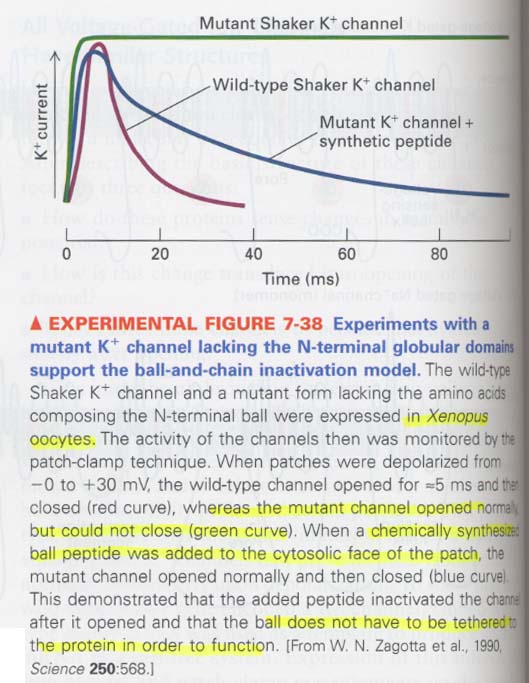
(7) Channel inactivation in voltage-gated K+ channels
--> soon after opening, spontaneously closing the channels
K+ channel; 4 α subunits + 4 β subunits
** Ball and chain model for inactivation
--> balls; positive charged, N-terminals
--> 아래 그림; blocking한 상태
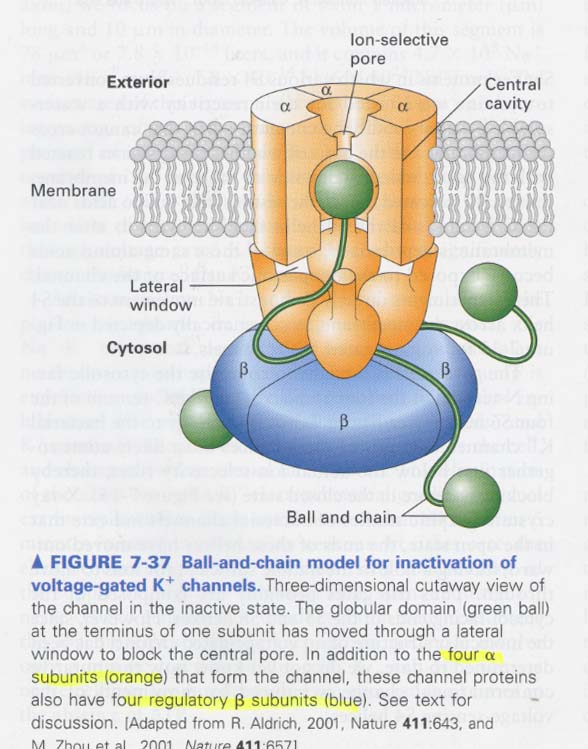
** experiment for demonstrating the ball domains as an inactivating segment
--> mutant; lacking the ball domain
--> in connecting chain experiment; shorter makes more rapid in the activation
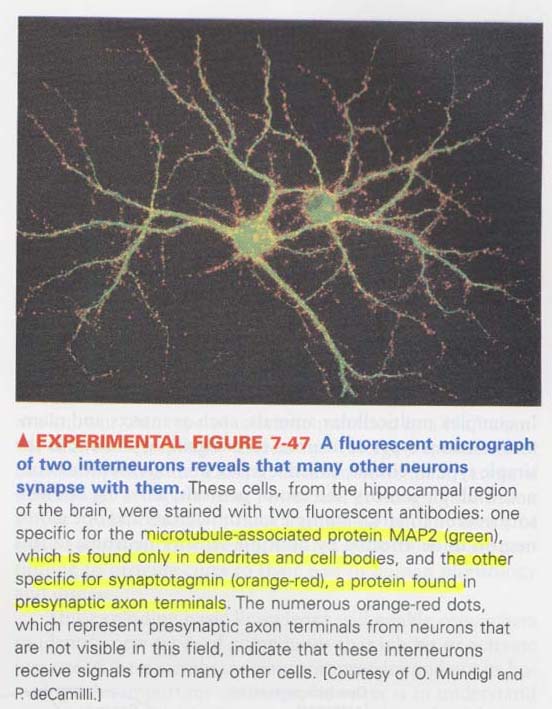
(8) How nerve cells make an action potential ?
** postsynaptic neurons receive signals from many presynaptic neurons
green (postsynaptic), orange-red (presynaptic)
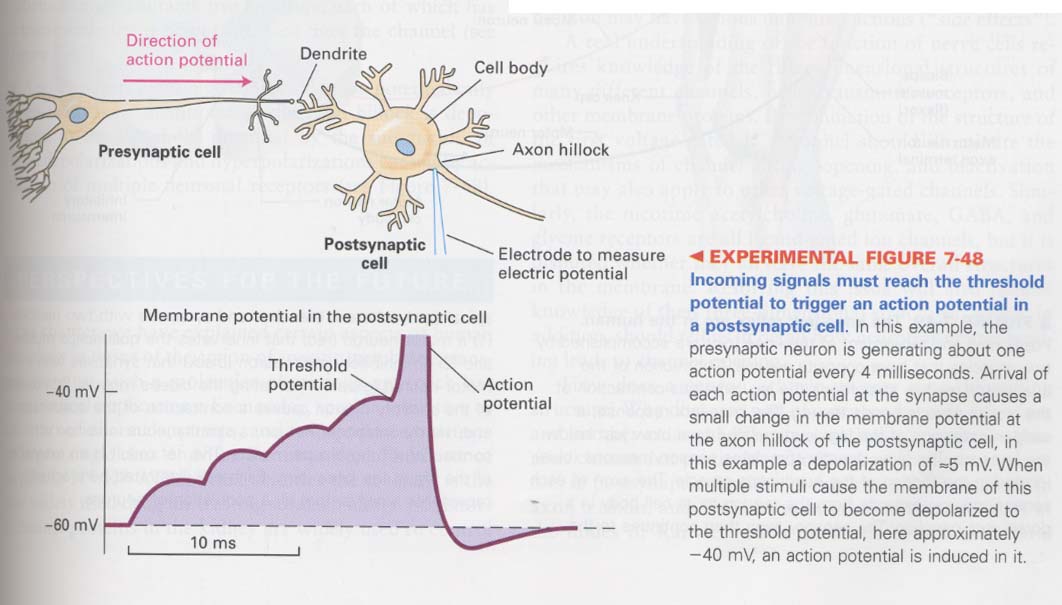
** generation of action potentials
--> combine of excitatory receptors with inhibitory receptors --> into axon hillock (summed together)
--> generate action potentials through threshold potential (all or nothing fashion)
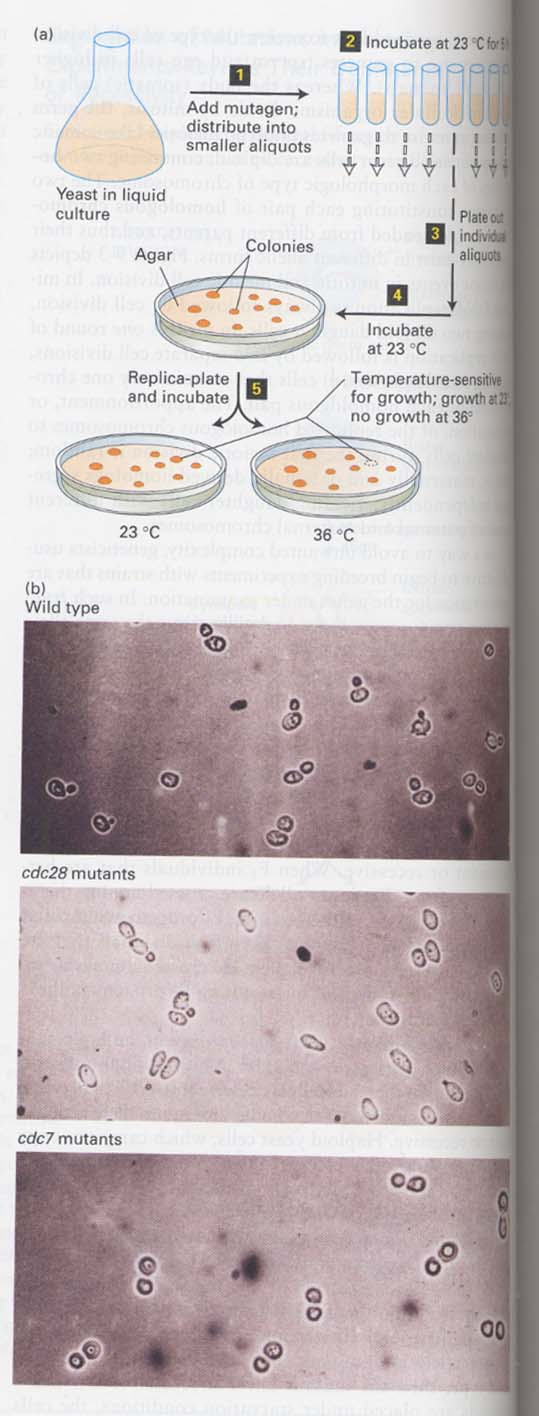
9장; Molecular genetic techniques and Genomics
(1) How do we study the essential genes in yeast ?
--> conditional mutation; in haploid cells
Tem. sensitive mutants 이용 (23oC 자라고, 36oC 안자람)
To isolate the genes involved in cell cycle
(2) complementation test
--> to see whether different recessive mutations are in the same gene or not
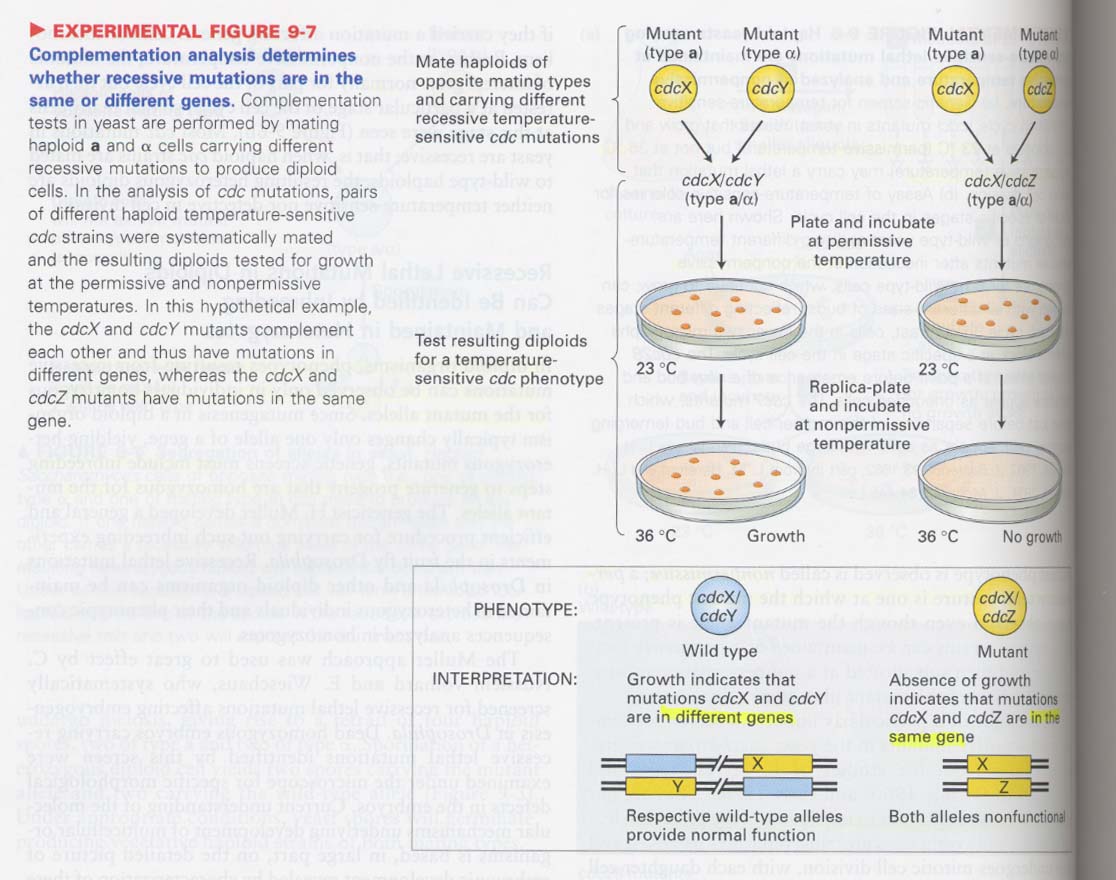
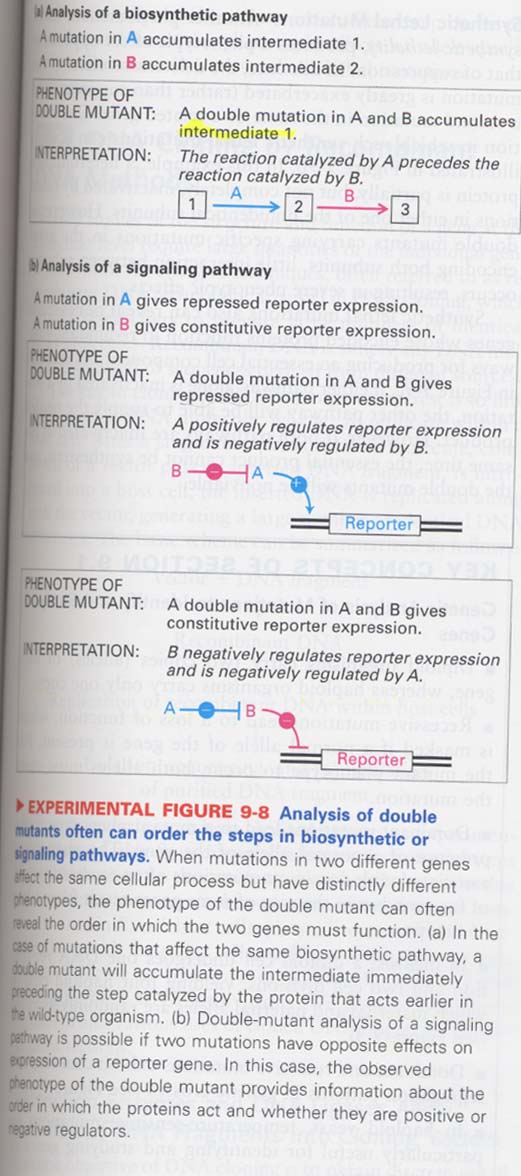
(3) double mutants 이용 (I)
--> to deduce the order in which proteins function
① biosynthetic pathway
② signaling pathway
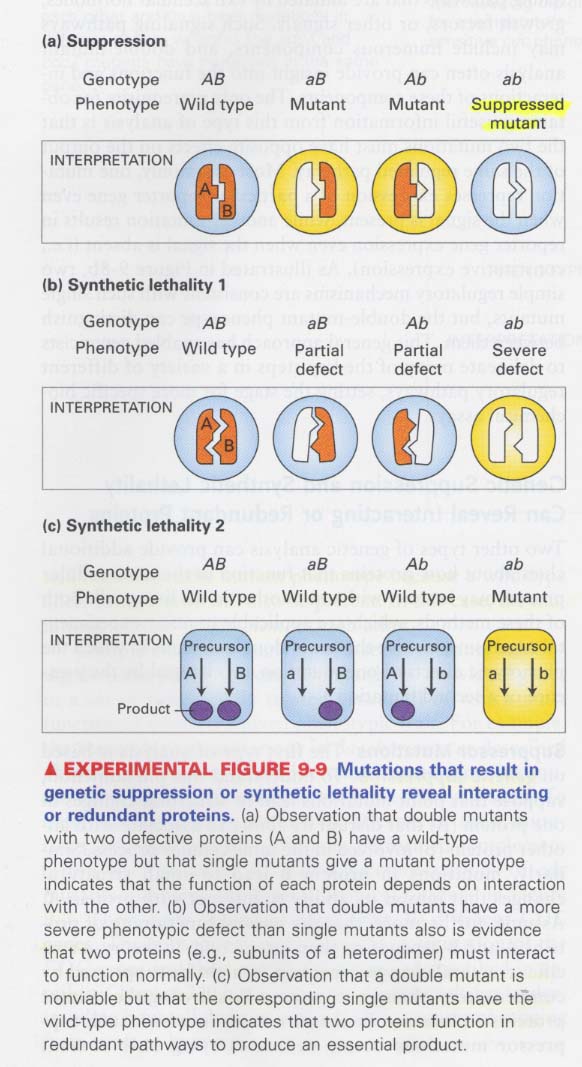
(4) double mutants 이용 (II)
--> to see how proteins interact with one another
① suppressor mutation; 다른 한쪽에서의 변이가 처음의 변이를 억제하는 효과
② synthetic lethal mutation; 다른 한쪽에서의 변이가 more severe효과를 주는 경우
어느한쪽에서의 변이가 효과가 없는 경우 (redundancy)
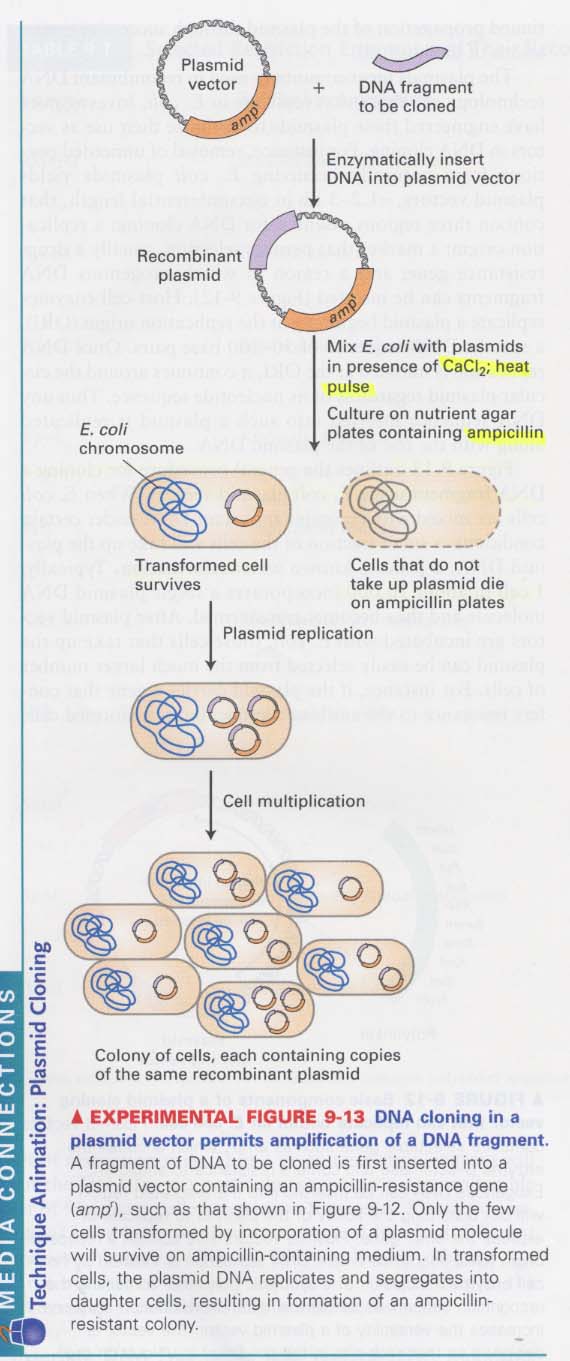
(5) Plasmid vector를 이용한 DNA cloning 원리
(6) λ phage vector를 이용한 cDNA library 제조
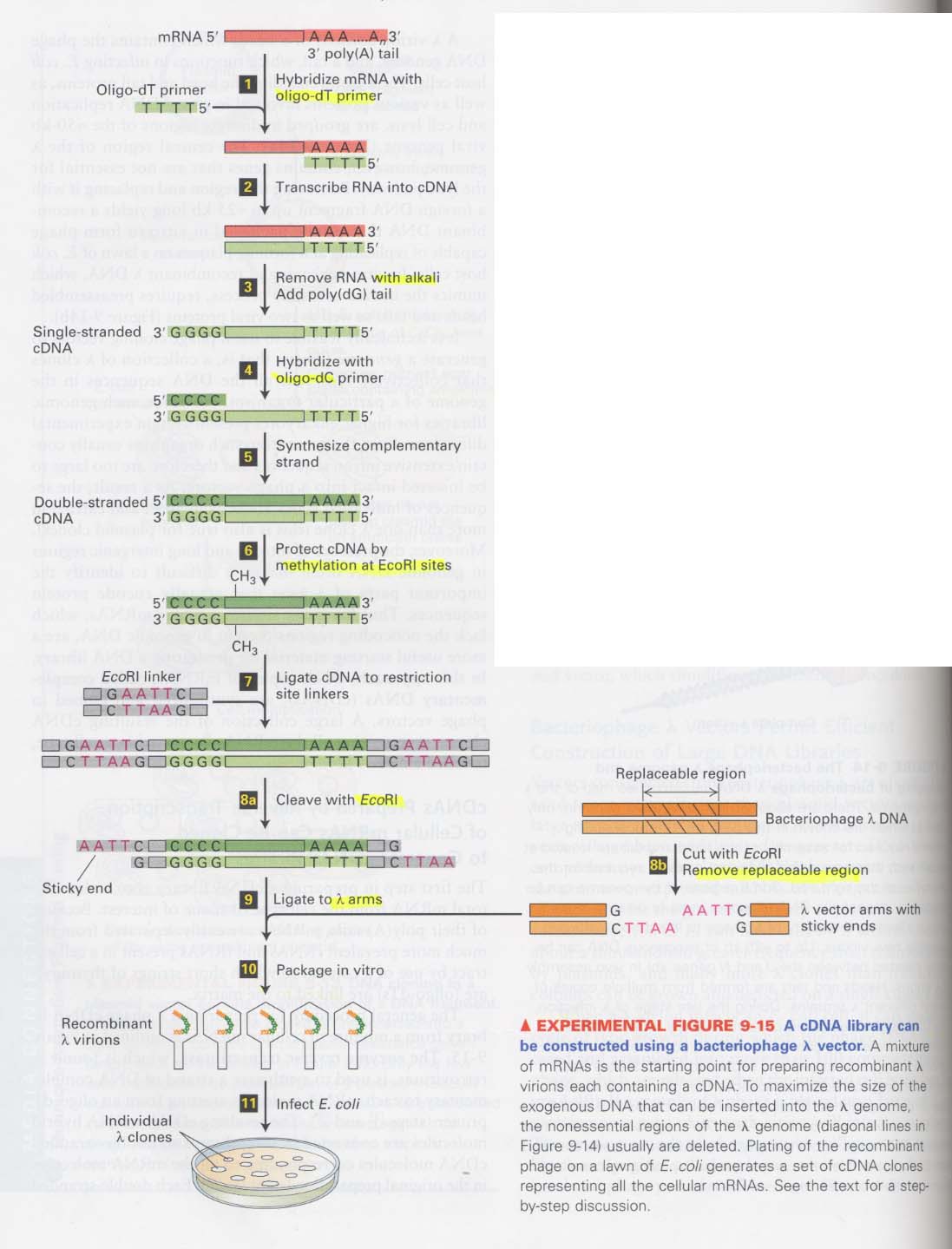
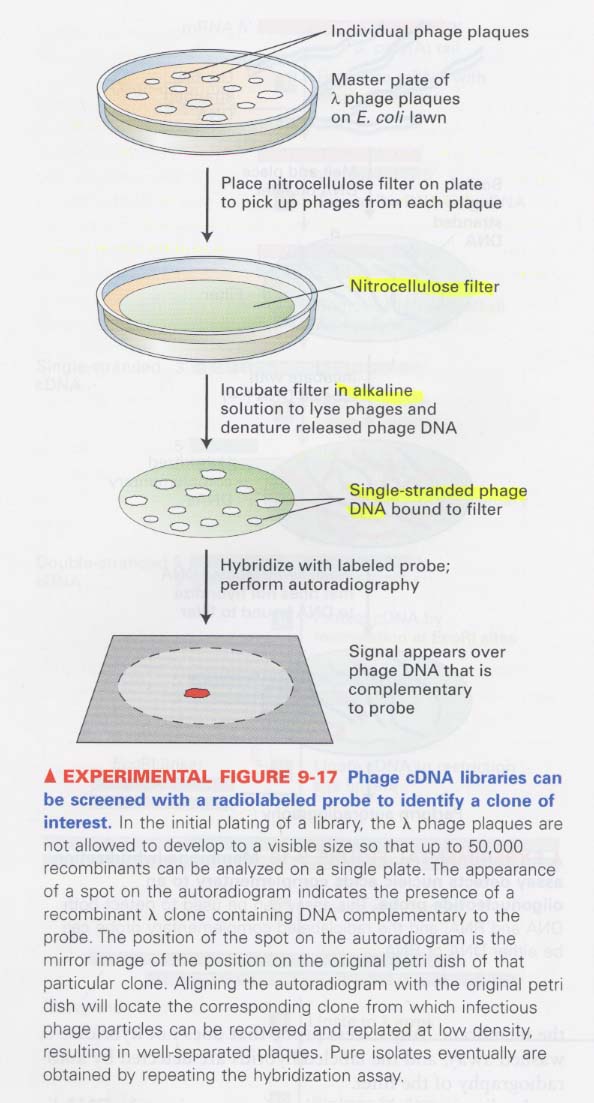
(7) λ phage vector를 이용한 cDNA library 로부터 interest clone을 조사하는 방법
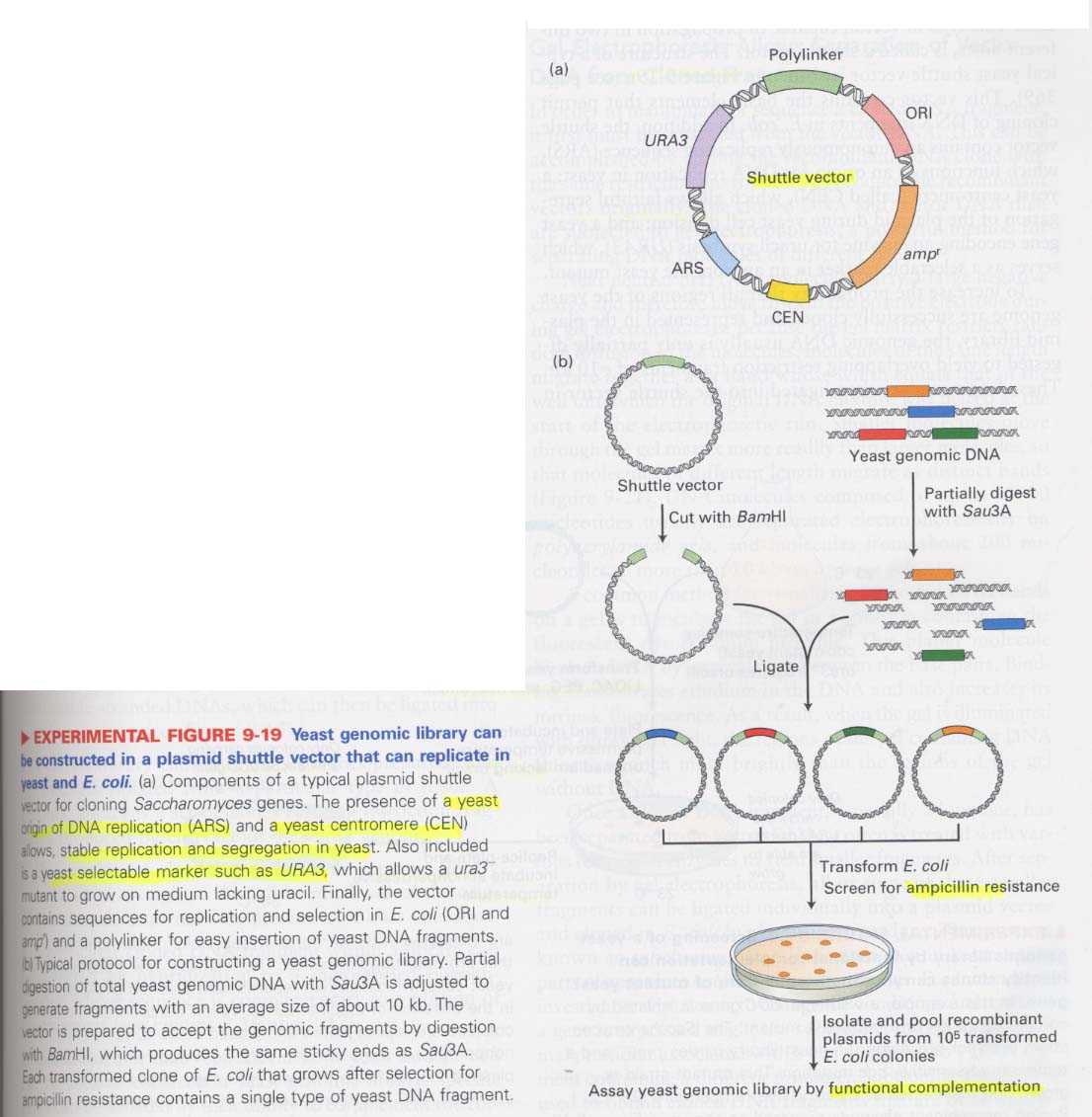
(8) shuttle vector를 이용한 yeast genomic library 제조
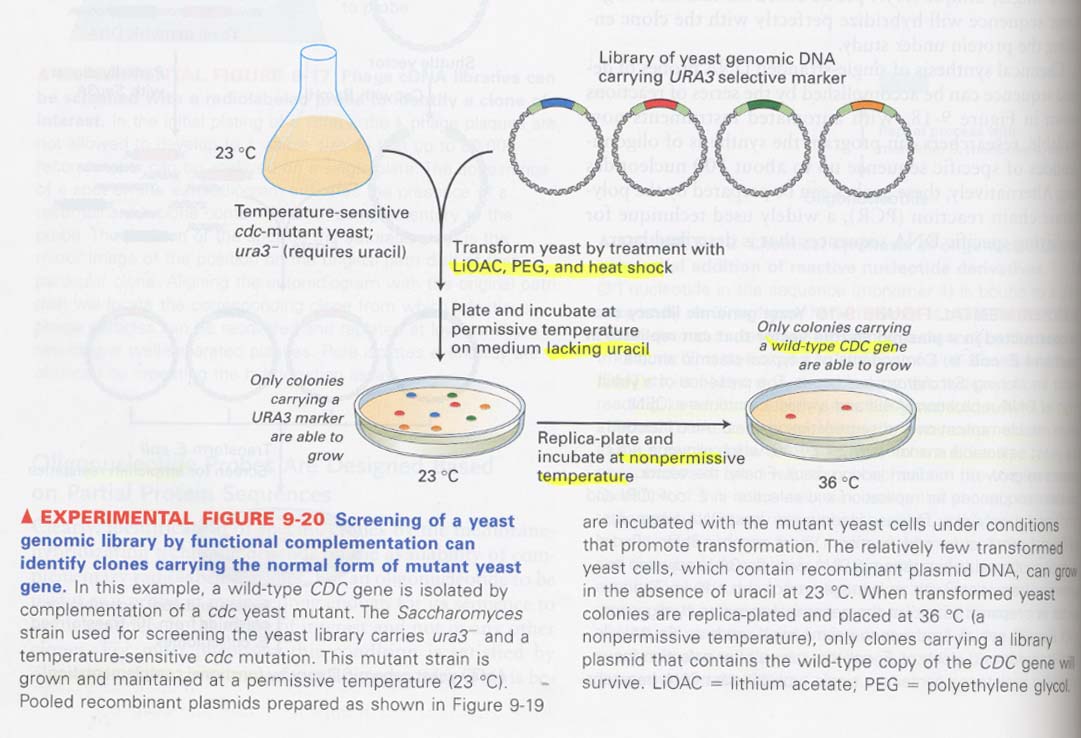
(9) yeast genomic library를 이용한 functional complementation
--> mutation을 야기한 유전자의 분석에 많이 이용
클로닝한 DNA fragment의 분석;
(10) electrophoresis를 통한 DNA size별 구분
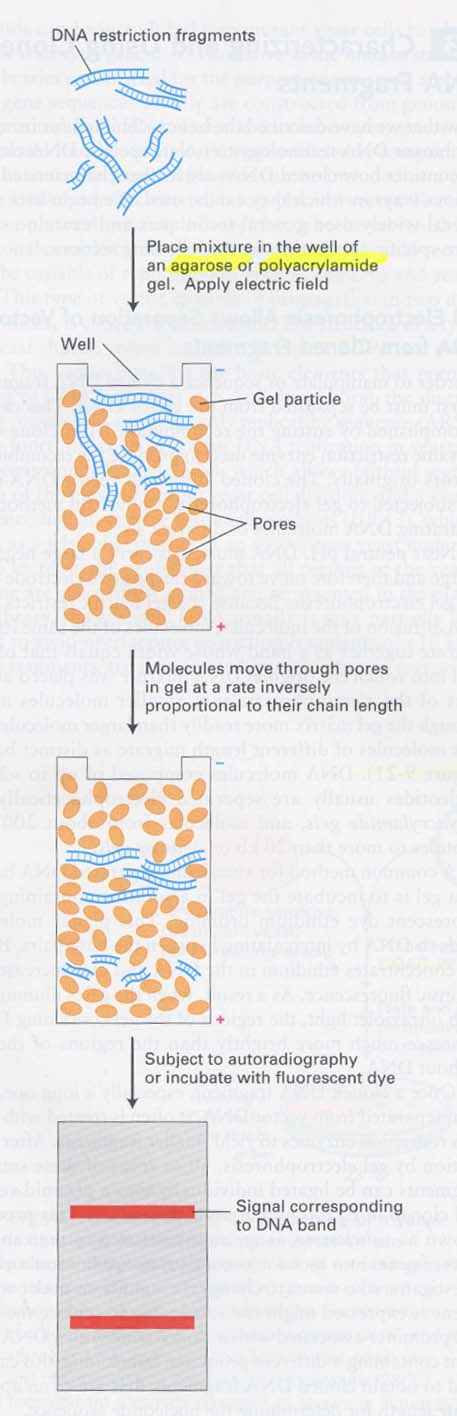
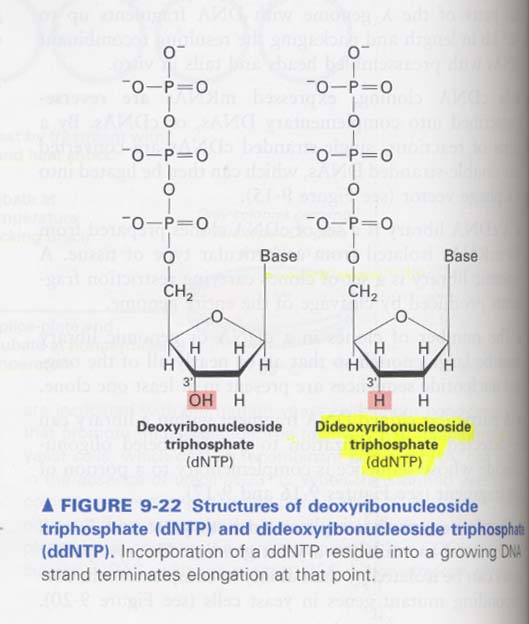
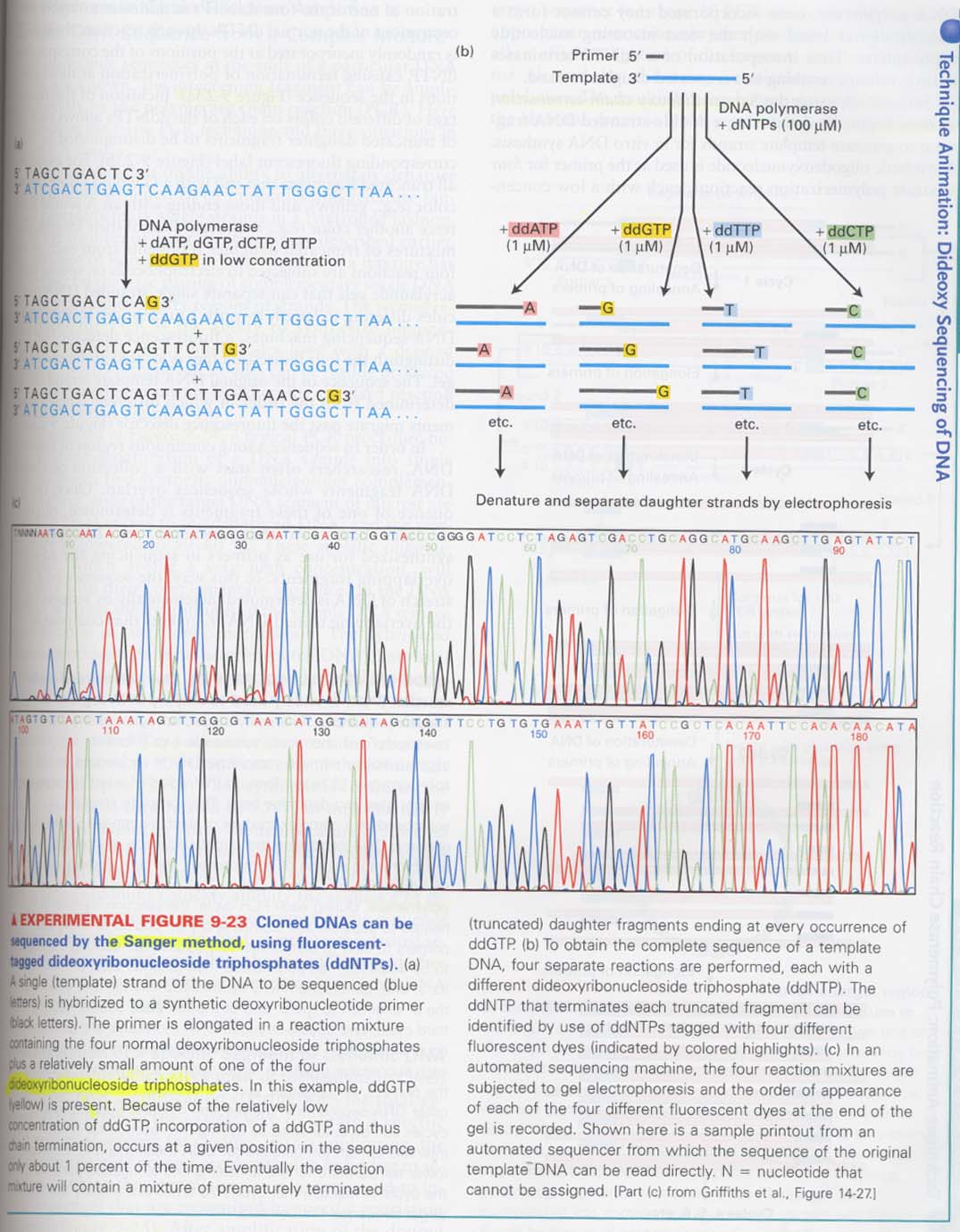
(11) DNA sequencing; Sanger method
--> termination에 사용되는 dideoxyribonucleoside triphosphate의 구조
--> sequencing의 원리 및 결과
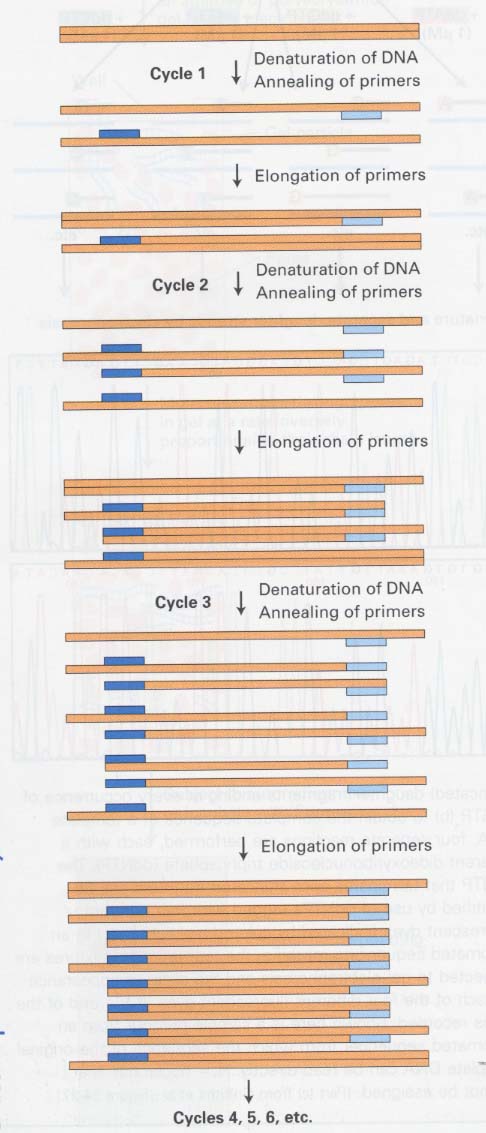
(12) PCR (polymerase chain reaction)
--> a million-fold after 20 cycles
(13) target gene cloning by PCR
--> primer 제조시 enzyme site를 삽입, 10kb 이상의 fragment를 주입할 수 있음
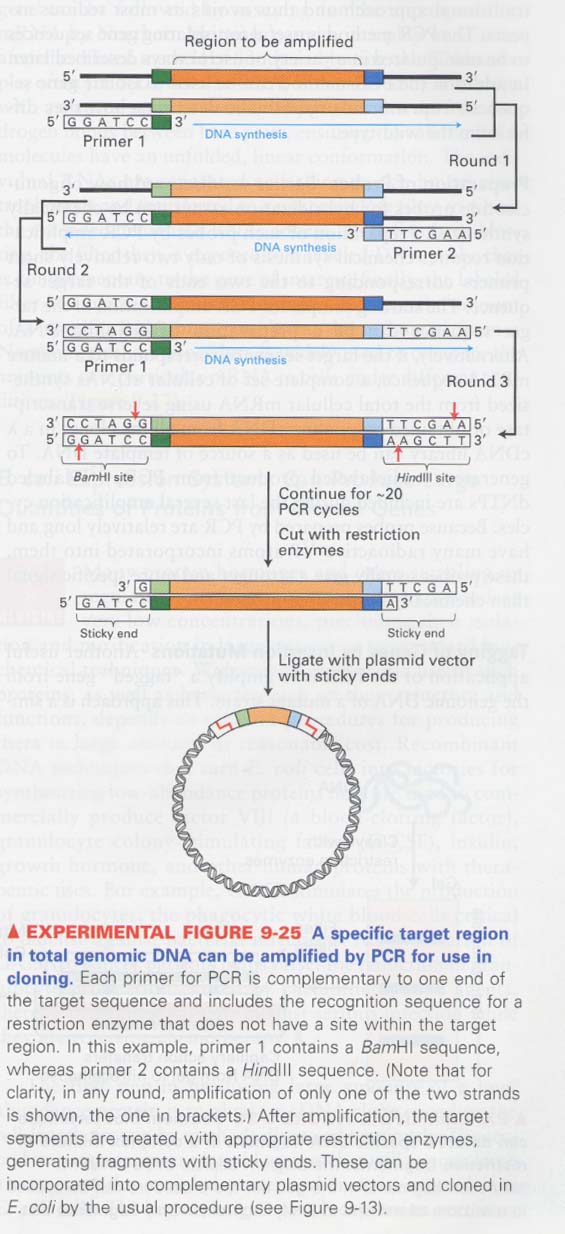
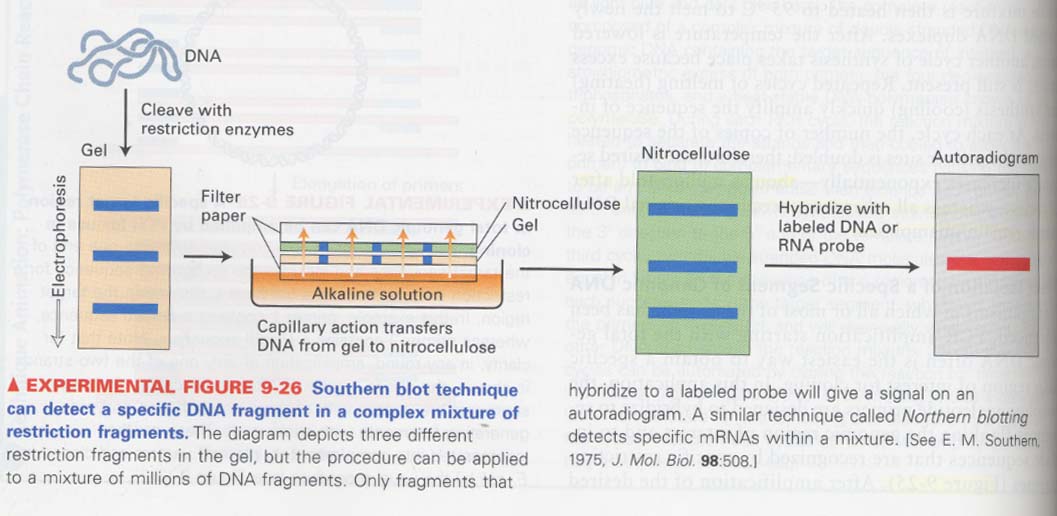
(14) southern blot
--> detection of single specific DNA from DNA mixture
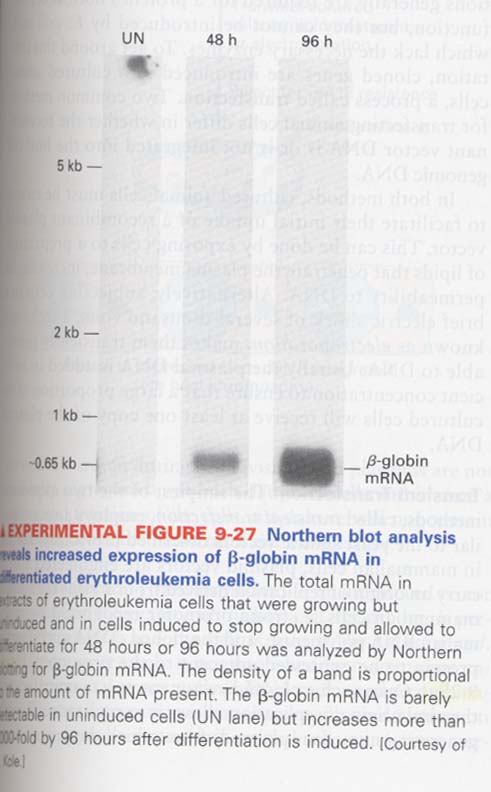
(15) northern blot
--> detection of single specific RNA from RNA mixture
--> ex) 적혈종양세포 분화과정에 있어서 β-globin mRNA 발현분석
(16) E. coli expression system
--> recombinant protein의 제조, for production of a large amount of protein
ex) G-CSF, factor VIII, insulin, growth hormone
--> lac promoter 및 IPTG 이용

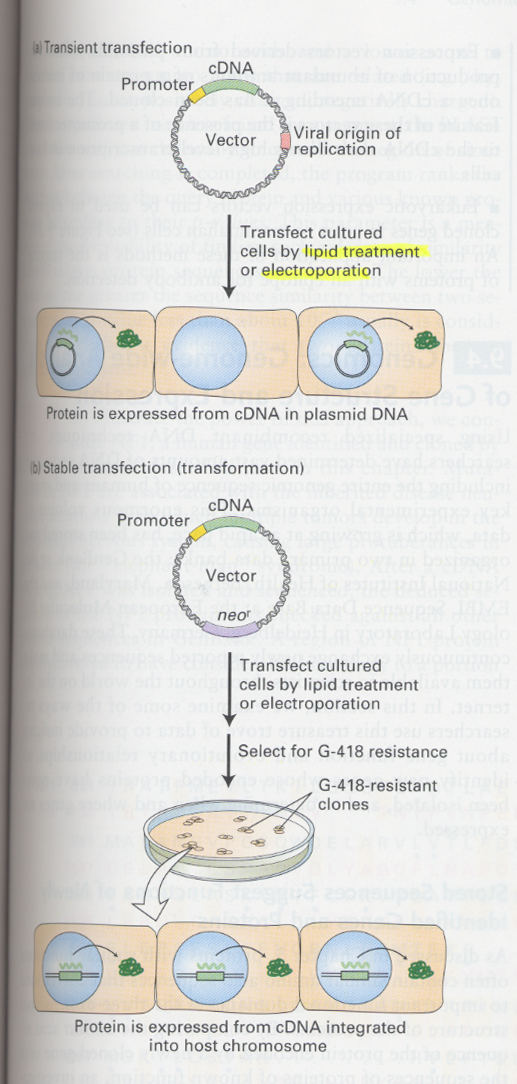
(17) Mammalian cell expression system
--> to overcome the post-translational modifications (glycosylation, hydroxylation)
--> by lipid or electroporation
--> neor ; neomycin phosphotransferase
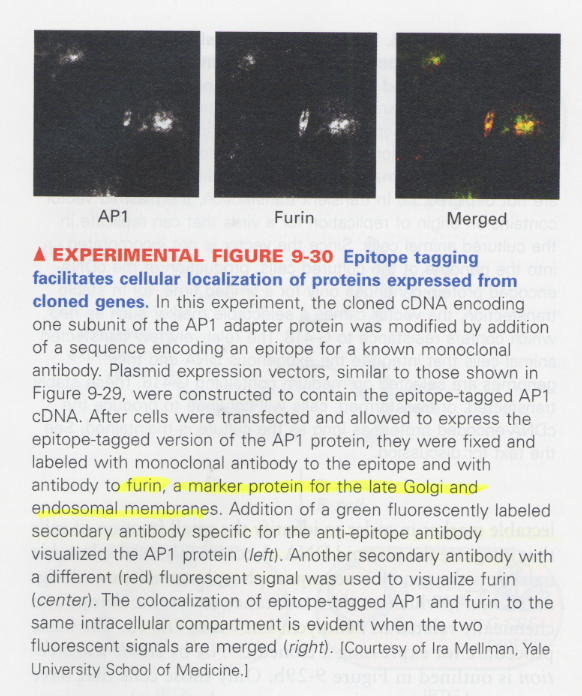
(18) epitope tagging
--> add a short Aas recognized by monoclonal antibody against the short fragment
--> 용도; to study the intracellular locolization of proteins
ex) AP1 adaptor protein (involved in clathrin-coated vesicle formation)
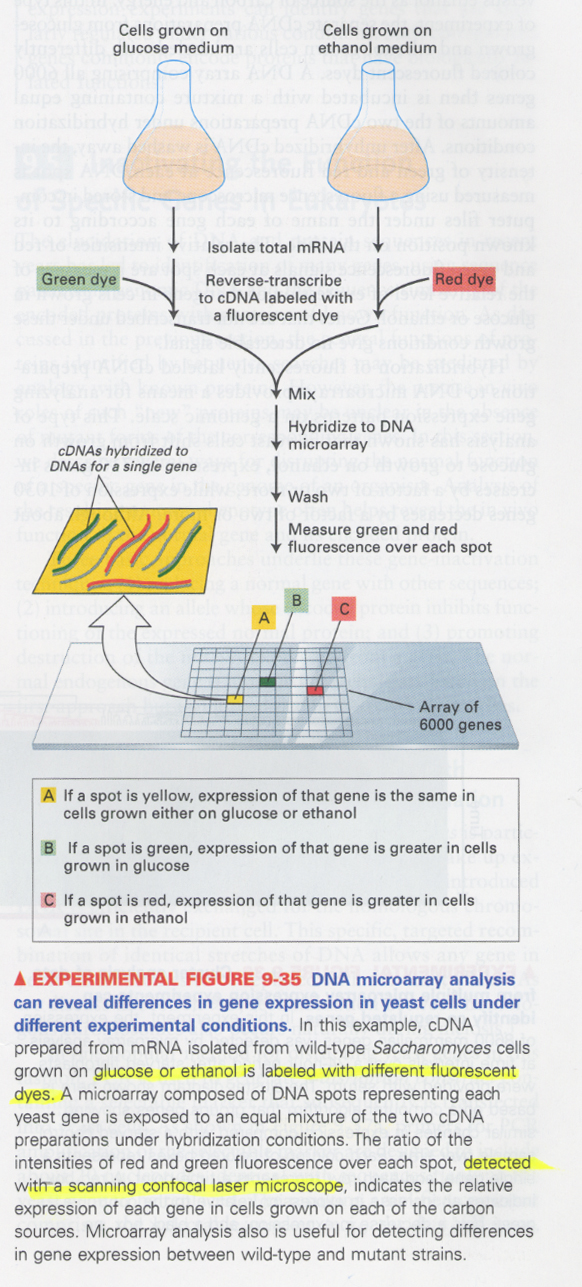
(19) microarray DNA
--> to see the gene expression pattern during specific physiological responses or developmental processes
--> ① DNA microarray; ~1kb coding fragment/spot
② DNA chip; ~20bp oligonucleotide/spot
ex) glucose or ethanol하에서 발현되는 효모유전자 pattern 분석
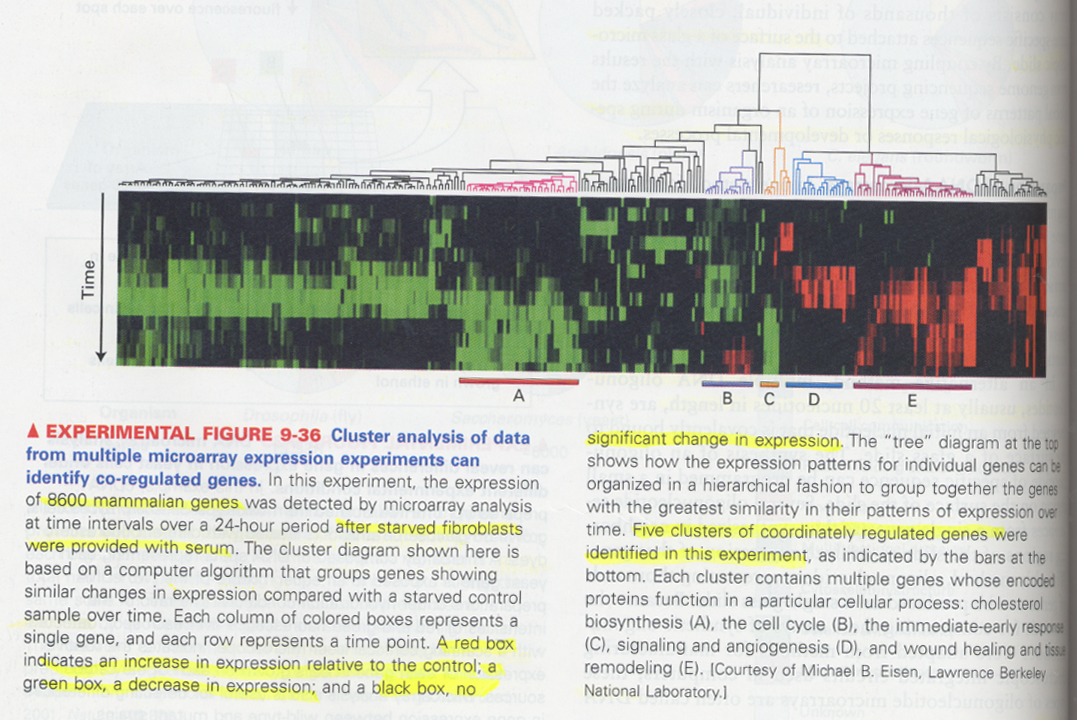
(20) cluster analysis
--> genes showing a similar gene expression in a single DNA microarray can be different in the biological function
--> to examine the closely related, co-regulated gene expression
(ex) check the expression pattern after serum addition
--> red; incease, green; decrease, black; no change
--> cholesterol synthesis, cell cycle, immediate-early genes, signaling and angiogenesis, wound healing
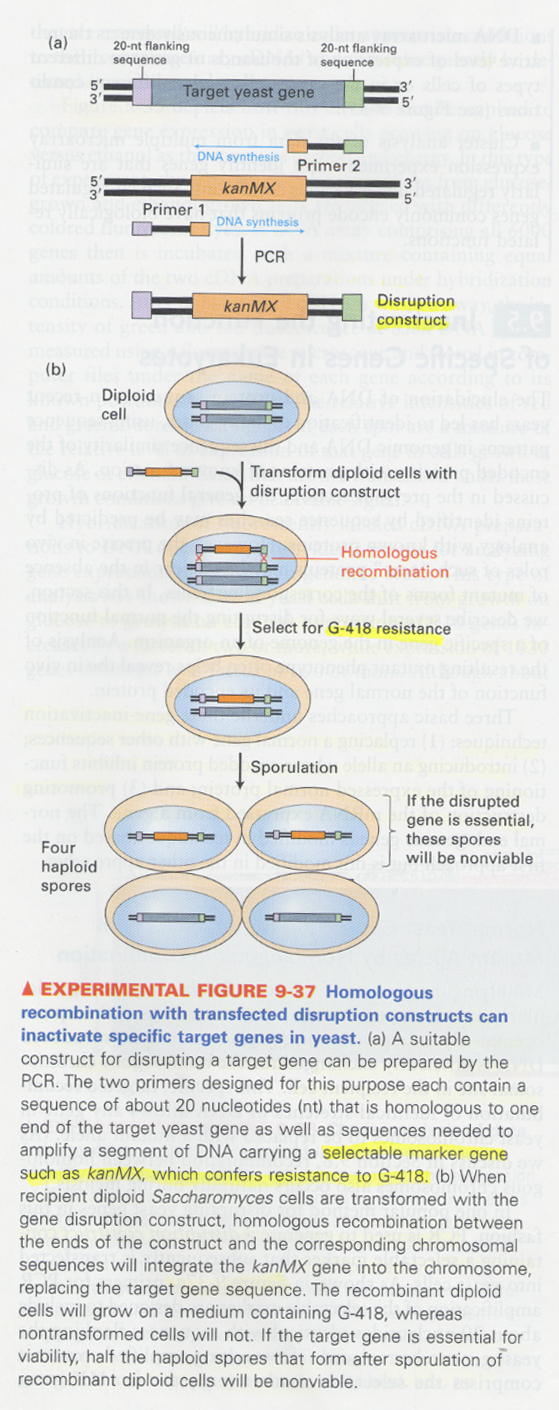
(21) homologous recombination in yeast
--> primers; use of flanking sequences of target sequence
--> selection; kanMX (resistance for G418)
(22) gene-knockout in mice
① knockout mutation in ES cells
--> two selectable markers; neo (resistance for G418), tk (sensitive for ganciclovir)
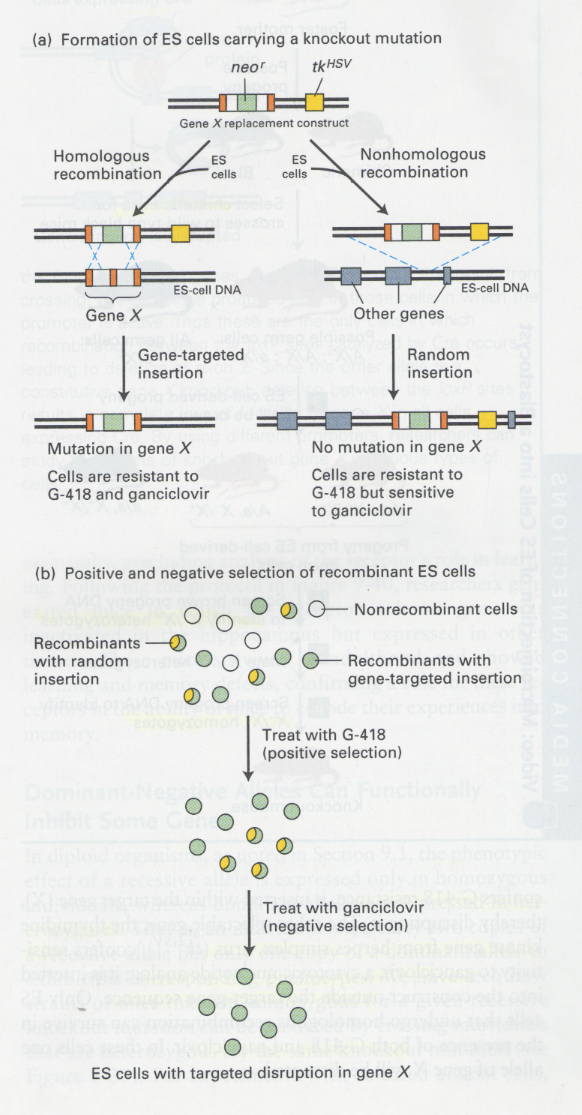
② production of knock-out mice
--> injection of ES cells (brown) into 4.5 days blastocyst (black)

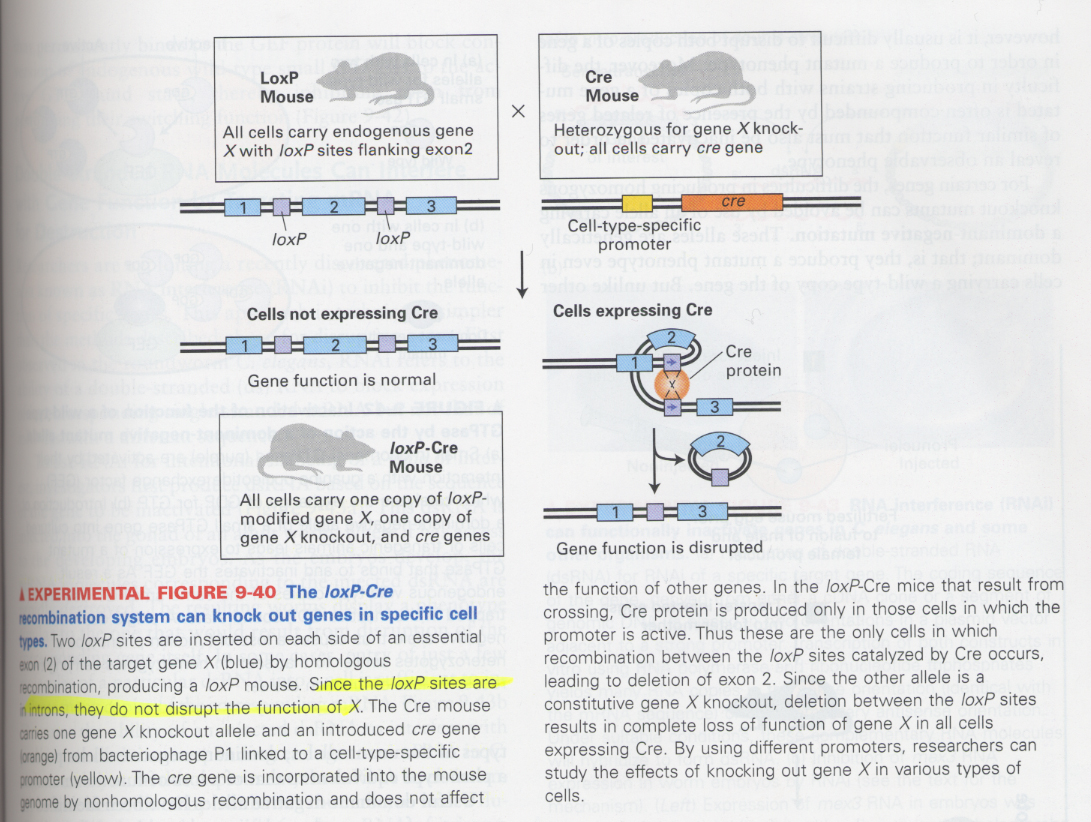
(23) gene-knockout in specific tissues
--> to examine the effect of knock-out mutation in the specific tissues or specific stage in development
--> loxP-Cre recombination technique; loxP (a site-specific recombination site), Cre (enzyme for recombination in loxP)
--> 용도; NMDA (N-methyl-D-aspartate) glutamate receptor in the hippocampus -- important in learning and memory
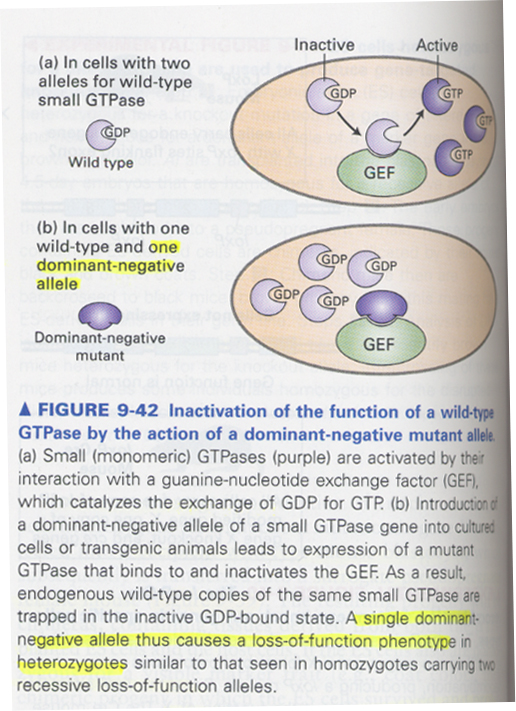
(24) dominant-negative mutation
--> 정의; cause a loss-of-function in heterozygotes
--> 장점; avoid the difficulty of obtaining the homozygous knockout mice
use of cultured animal cells
problem of related or similar functional genes
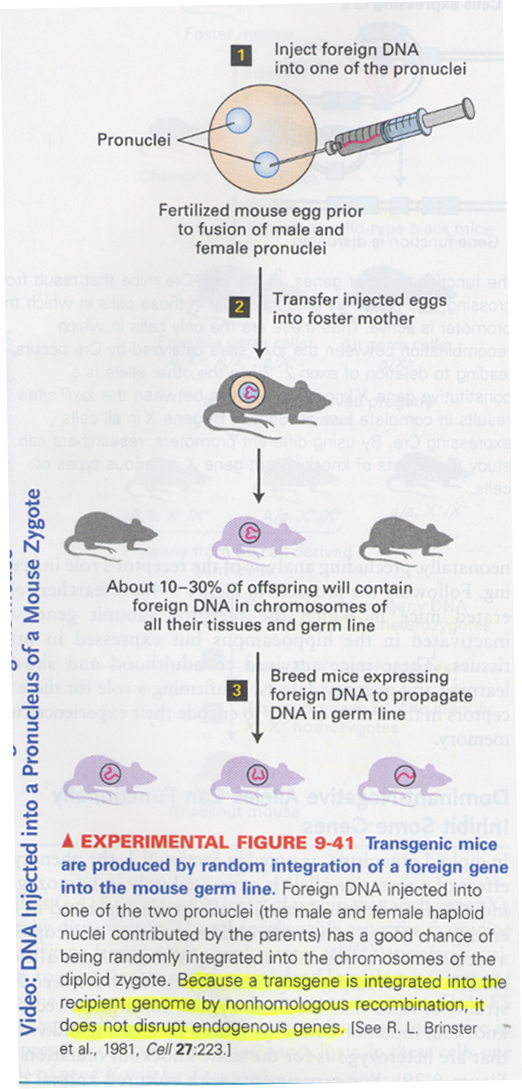
--> transgenic mice 제조; randomly inserted, nonhomologous recombination
regulated promoter
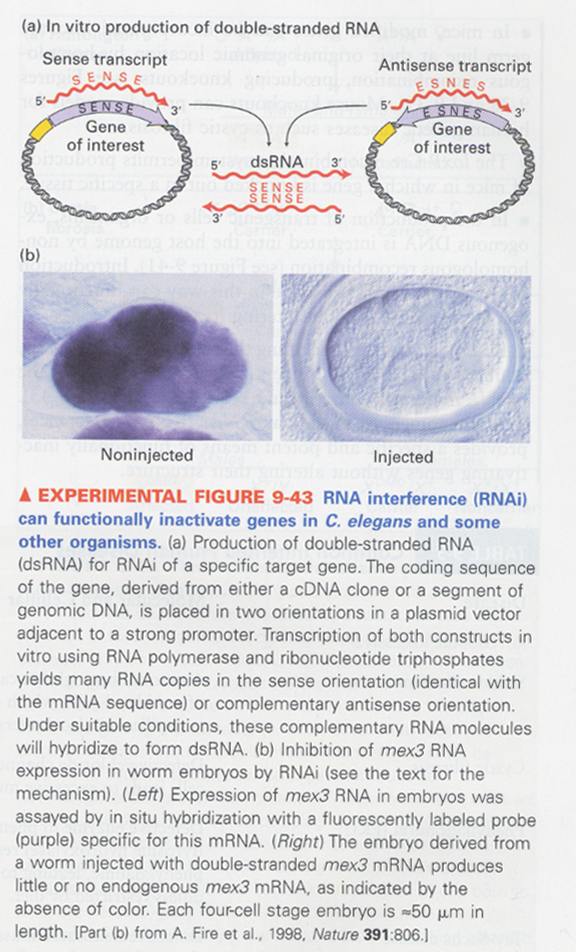
(25) RNA interference (RNAi)
--> for inactivating of a specific gene
--> use of double-strand RNA, detection through in situ hybridization
(ex) C. elegans, Drosophila, plants, zebrafish, spiders, Xenopus, mice
--> how; by specialized RNA-processing enzymes
function; defense against viruses, for regulation of certain endogenous genes
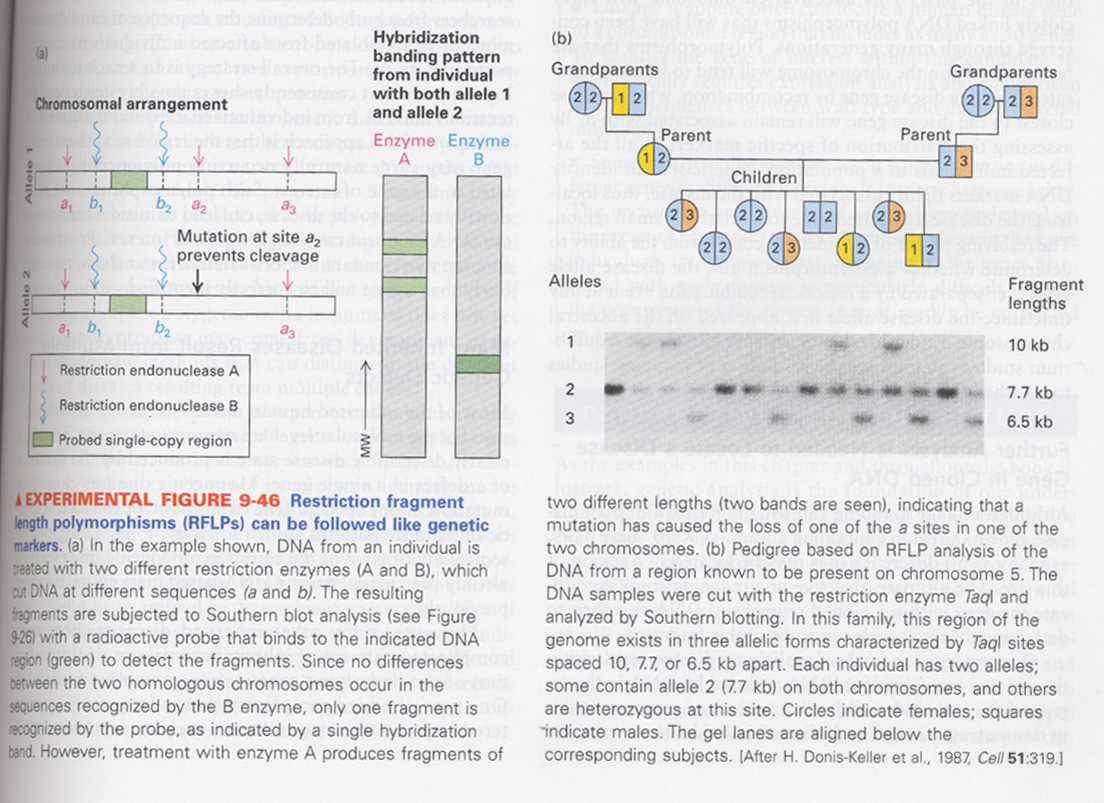
(26) DNA polymorphism
① Restriction fragment length polymorphism (RFLP)
--> 그림a; mutations cause to change a restriction enzyme site
--> pedigree 조사; linkage of a certain allele to the inherited trait or disease
② single nucleotide polymorphism (SNP)
③ simple sequence repeats (SSR, microsatellites); 1, 2, or 3 base sequence repeat number
--> detected by PCR analysis and DNA sequencing
10장; Molecular structure of genes and chromosomes
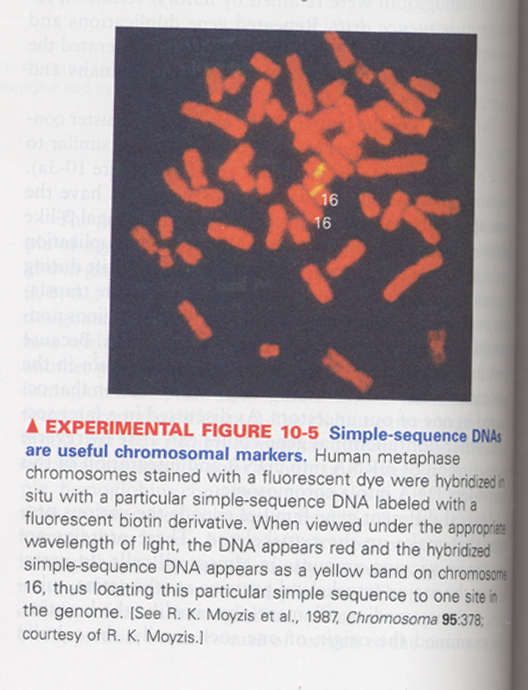
(1) Simple-sequence DNA (= satellite DNA) is located at the specific sites of chromosomes
--> fluorescence in situ hybridization (FISH)
--> satellite DNA; 14-500 bp, tandom repeats of 20-100 kb
--> centromere, telomere, chromosome arm
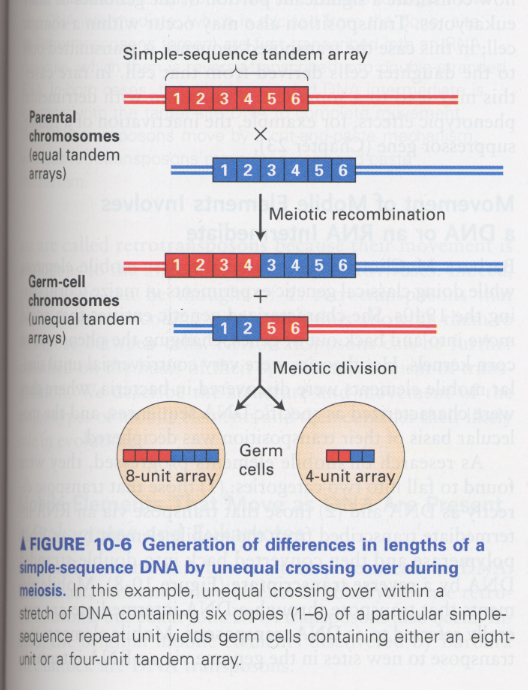
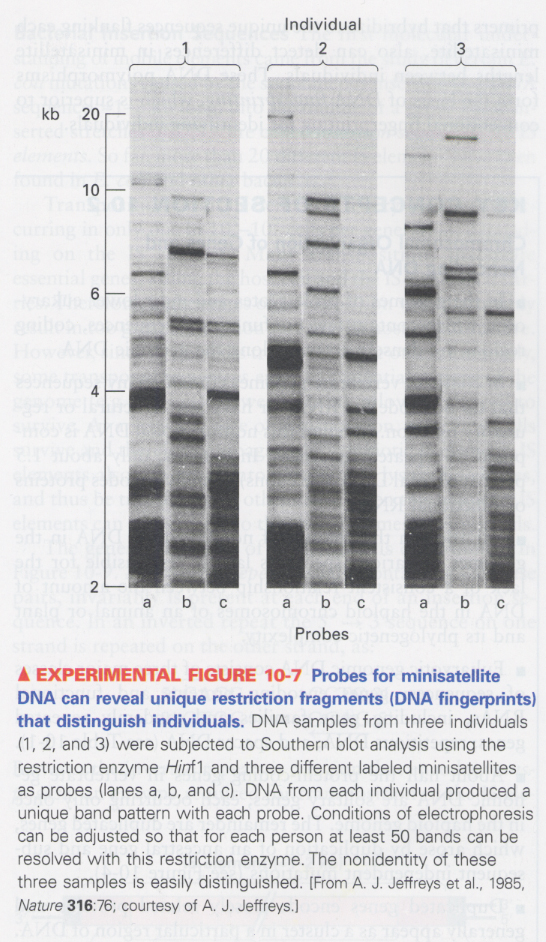
(2) DNA fingerprinting
--> repeat number of simple-sequence DNA is different among individuals
why; by unequal cossing-over (Fig 10-6)
--> minisatellite DNA; 15-100 bp, tandom repeats of 1-5 kb
① southern blotting; different minisatellite probes (a, b, c)
② PCR; primers for flanking sequences of minisatellites
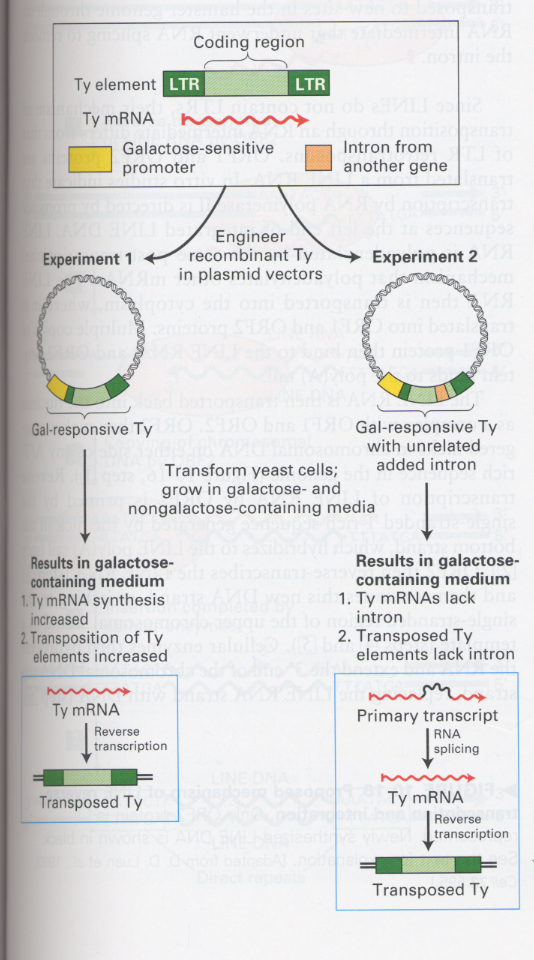
(3) Role of RNA intermediate in the transposition
--> in the LTR retrotransposon; reverse transcriptase, integrase
--> 오른쪽 panel; transposed된 Ty element에서 삽입된 intron을 볼 수 없음
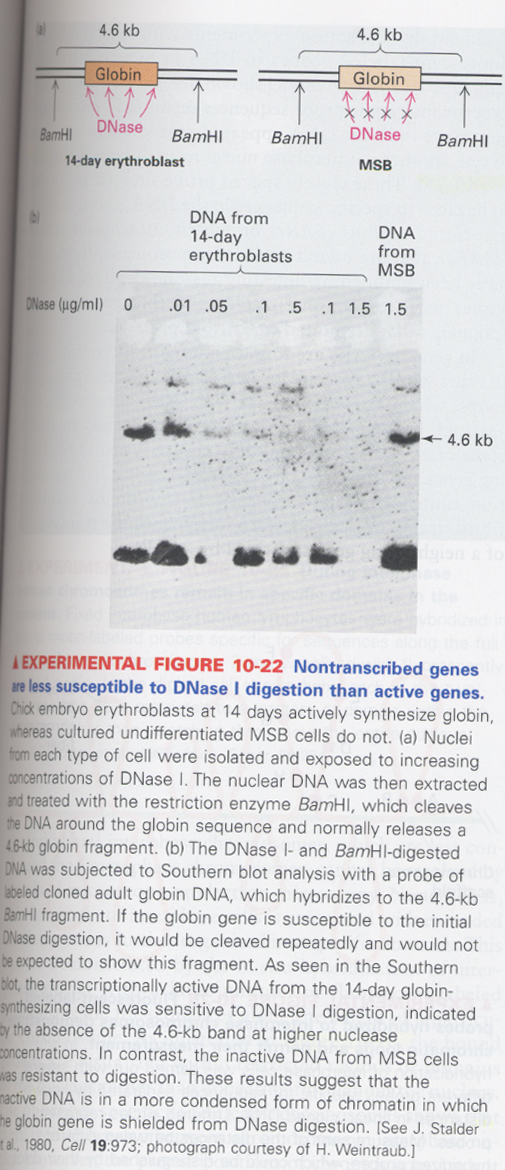
(4) chromatin condensation
--> transcribed gene is higher than the untranscribed one in histone acetylation
--> DNase sensitive
ex) erythrocyte is active in the globin gene expression
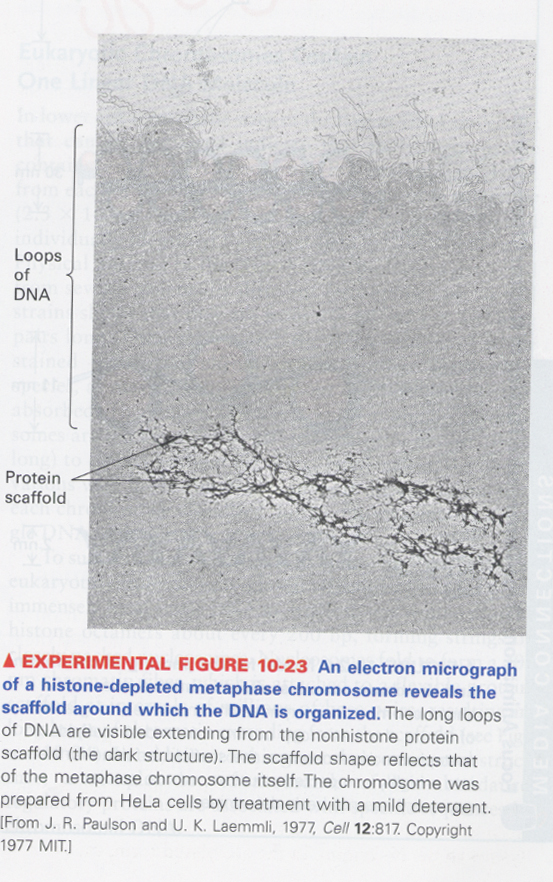
(5) chromosome scaffold
--> non-histone proteins also act as a chromosome scaffold in histone-depleted chromosomes
--> DNA loop structure in the chromosome
증거 (Fig10-25); A, B and C--> scaffold associated region (SAR)
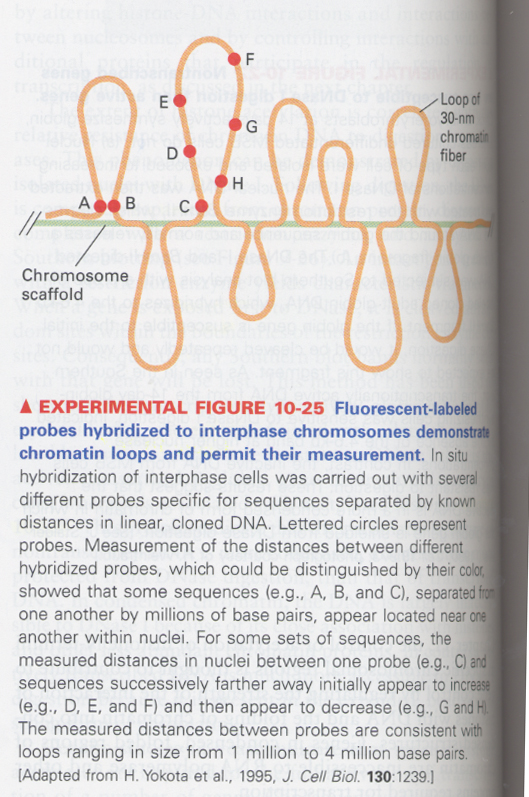
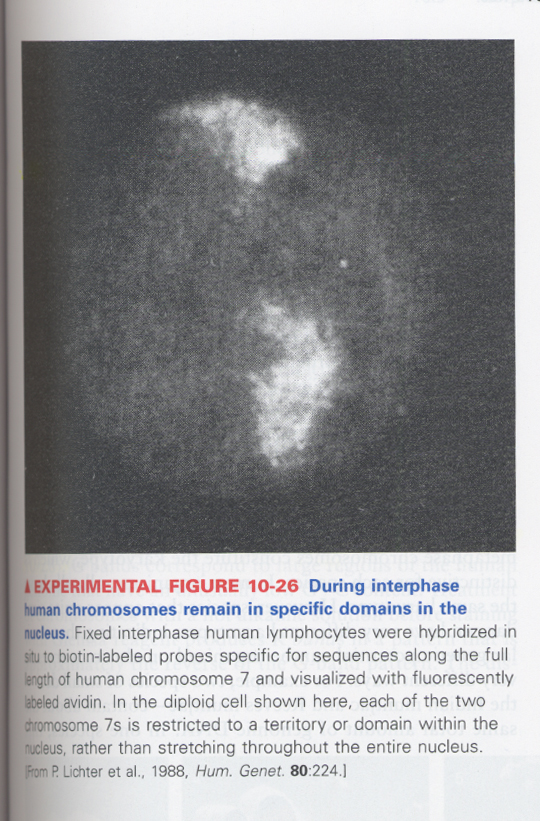
--> interphase chromosome is located within a specific, restricted regions of nucleus
ex) human chromosome 7
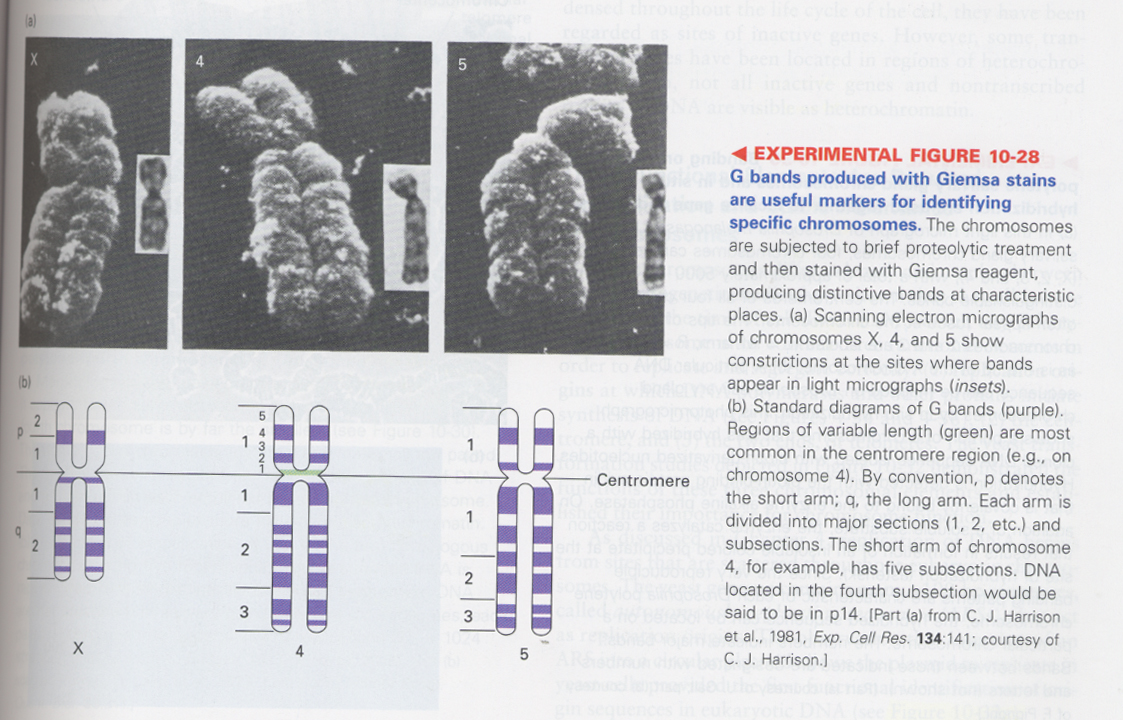
(6) chromosome band patterning
--> G bands; staining with Giemsa reagent after mild heat or proteolysis, detection of low G + C content
p; short arm, q; long arm
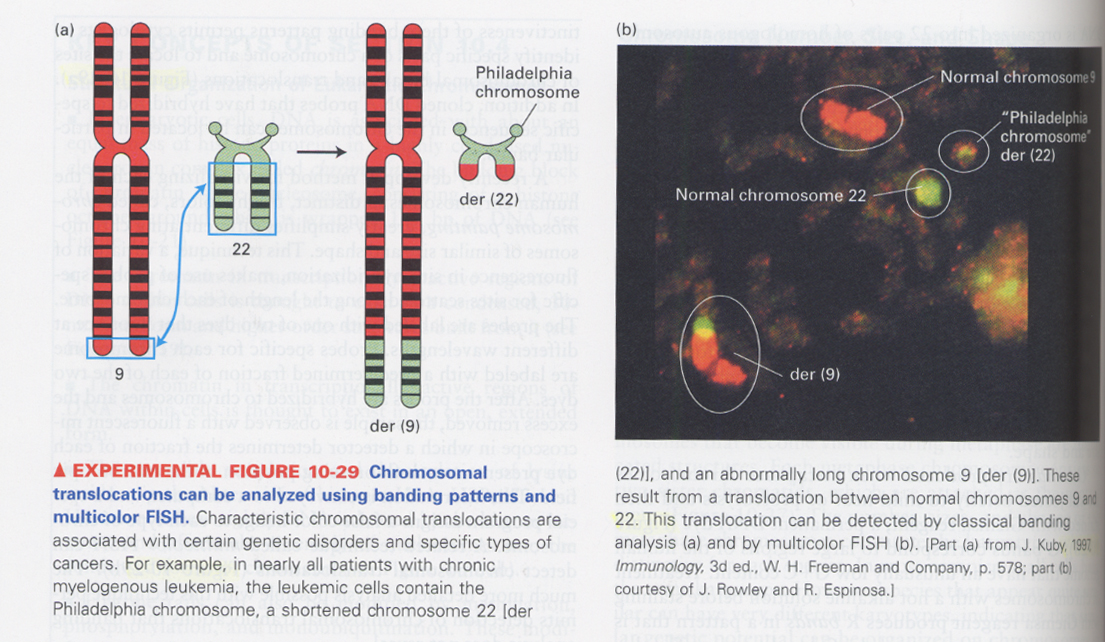
chromosome painting (multicolor FISH)
--> for detection of translocation that banding pattern analysis does not reveal the difference
ex) chronic myelogenous leukemia; philadelphia chromosome의 존재
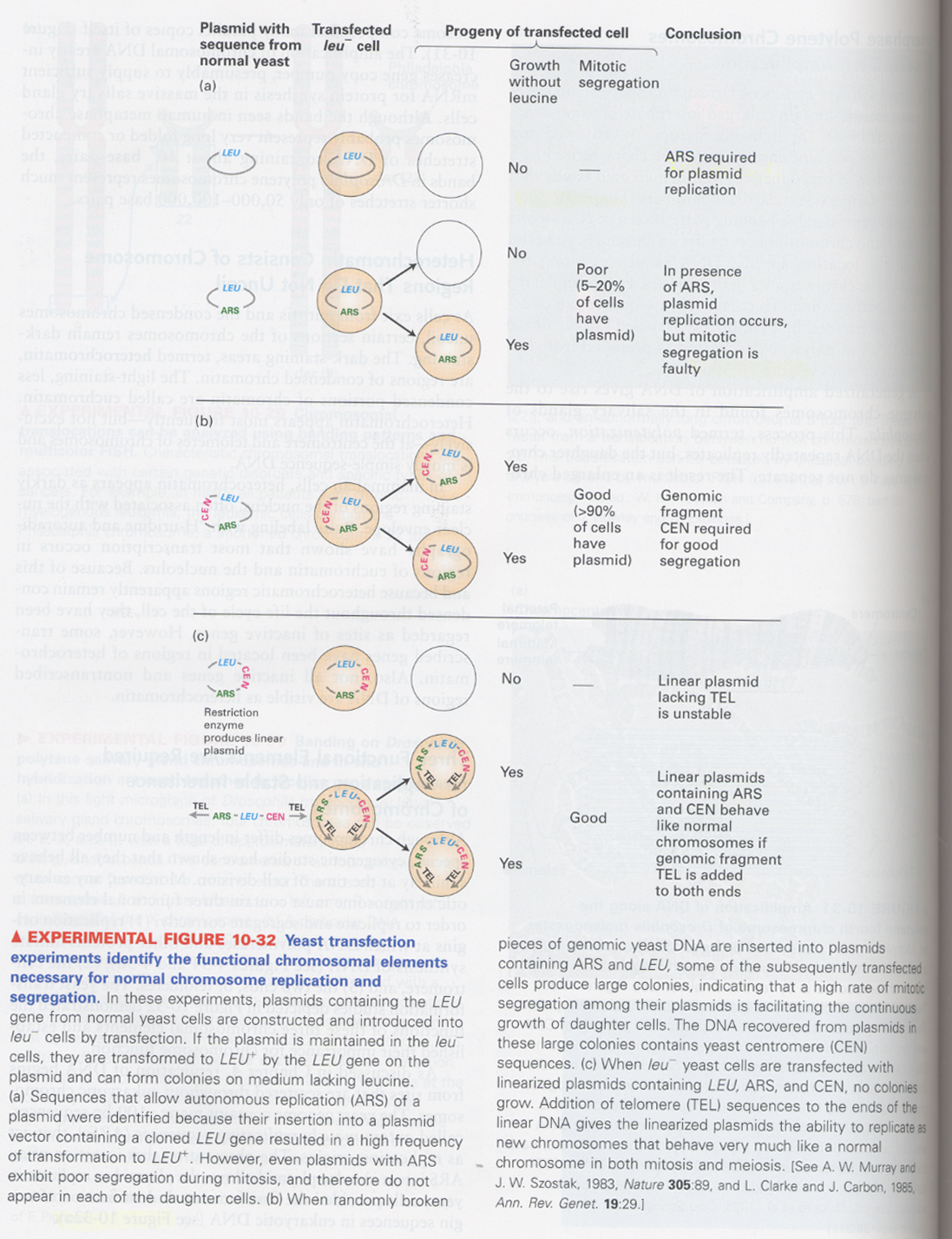
(7) important functional elements for chromosome replication and inheritance
① ARS (autonomously replicating sequence); replication origin
② CEN (centromere); for mitotic segregation
③ TEL (telomere); for mitotic segregation
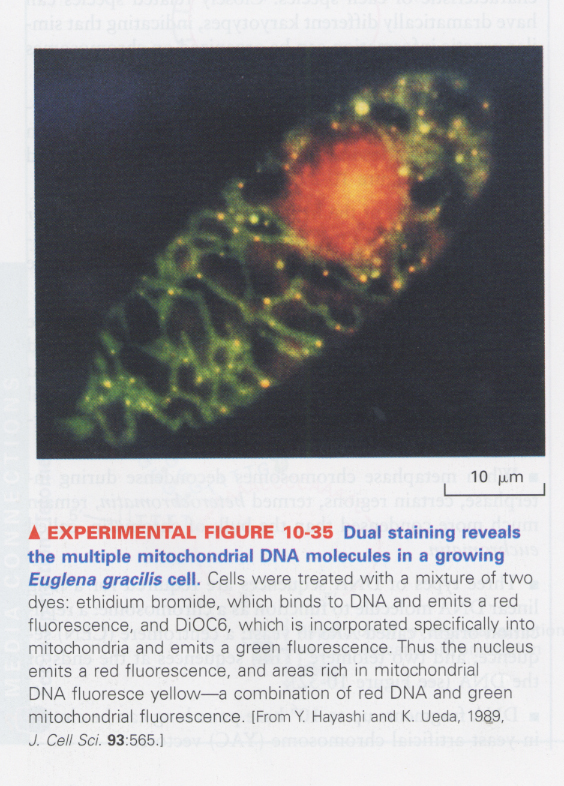
(8) mitochondria DNA detection
--> ethidium bromide (red), DiOC6 (green, for mitochondria)
--> yellow; indicates the mtDNA
11장; transcriptional control of gene expression
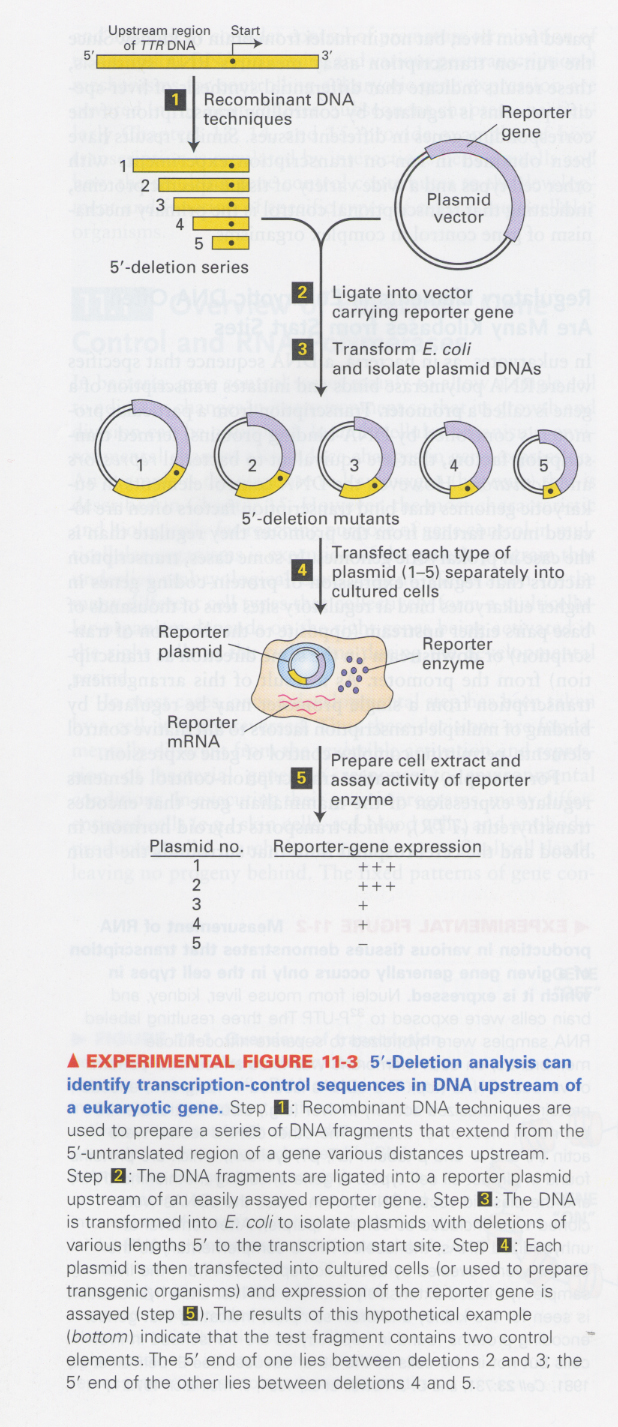
(1) characterization of transcription-control sequence of genes
--> 5' deletion experiment
--> reporter system; ① lacZ (β-galactosidase) ② luciferase ③ GFP
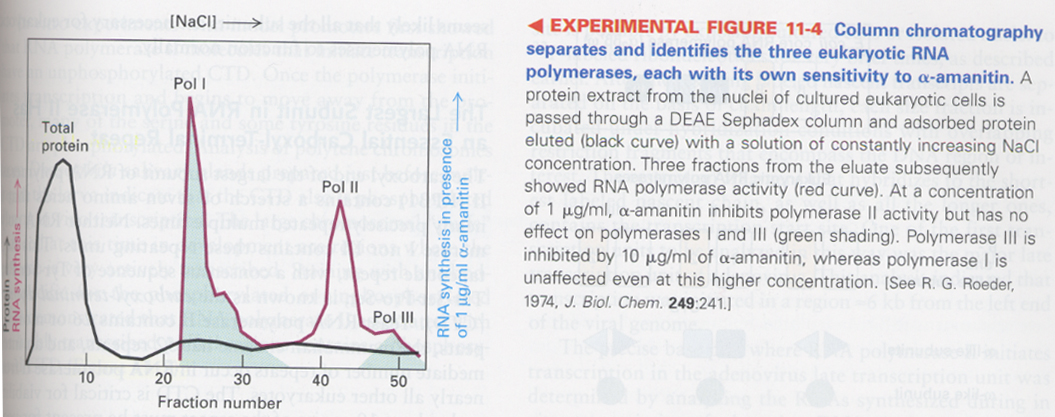
(2) separation of three kinds of eukaryotic RNA polymerases
--> diffrence in salt conc. in elutes, α-amanitin (cyclic peptide, 8Aas, mushroom) sensitivity; poly II is strong sensitive
(3) CTD (carboxyl-terminal domain) of eukaryotic RNA polymerase II
--> in the highly transcribed genes; phosphorylated (red)
--> CTD; heptapeptide (Tyr-Ser-Pro-Thr-Ser-Pro-Ser), important to viability
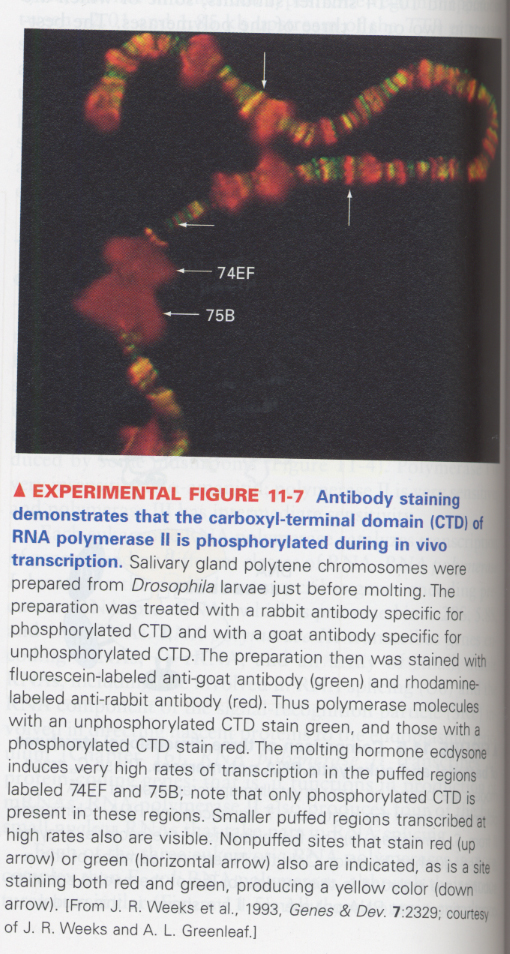
(4) Detection of an initiation site of RNA transcript
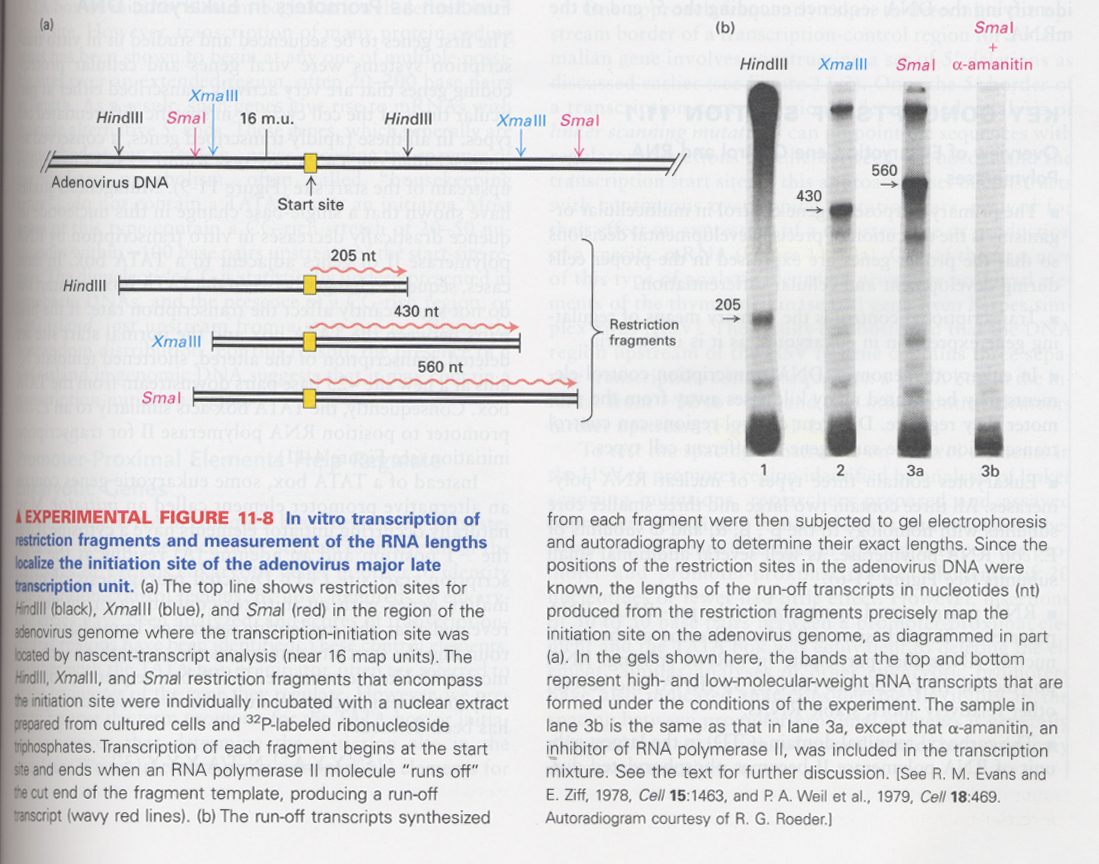
--> nuclear run-off assay in the presence of 32P-labeled ribonucleoside triphosphates
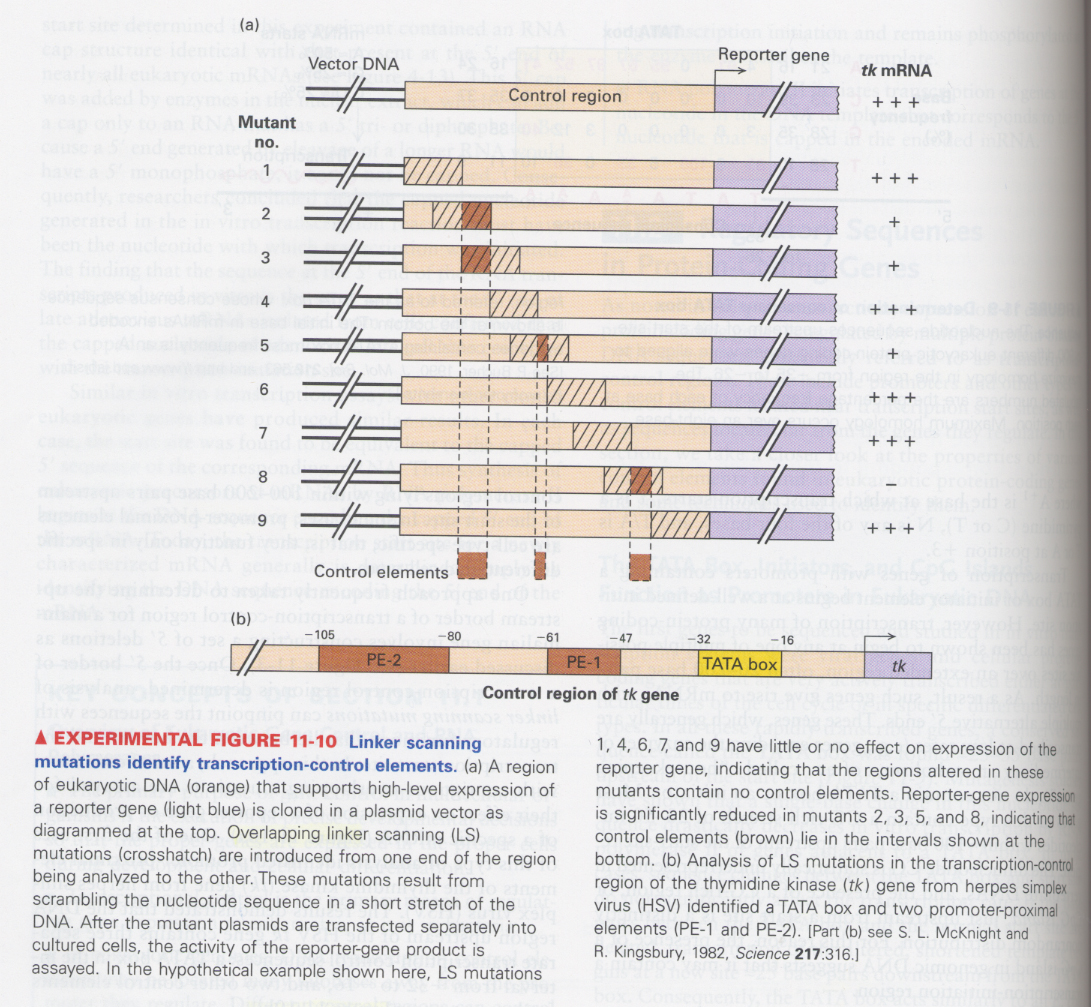
(5) Linker scanning mutation
--> to pinpoint the exact regulatory sequence (promoter-proximal elements) for gene expression
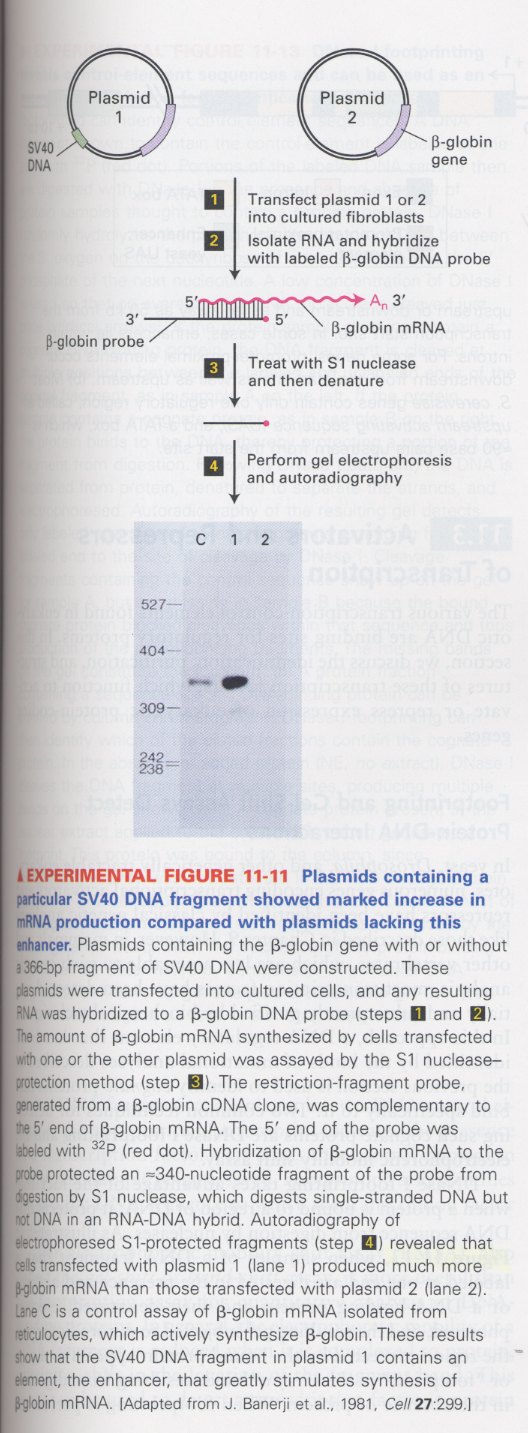
(6) Detection of enhancer (S1 nuclease protection assay)
--> SV40 DNA contains an enhancer for gene transcription
--> C (control); erythrocytes, 1 and 2; fibroblasts
(7) protein-DNA interaction
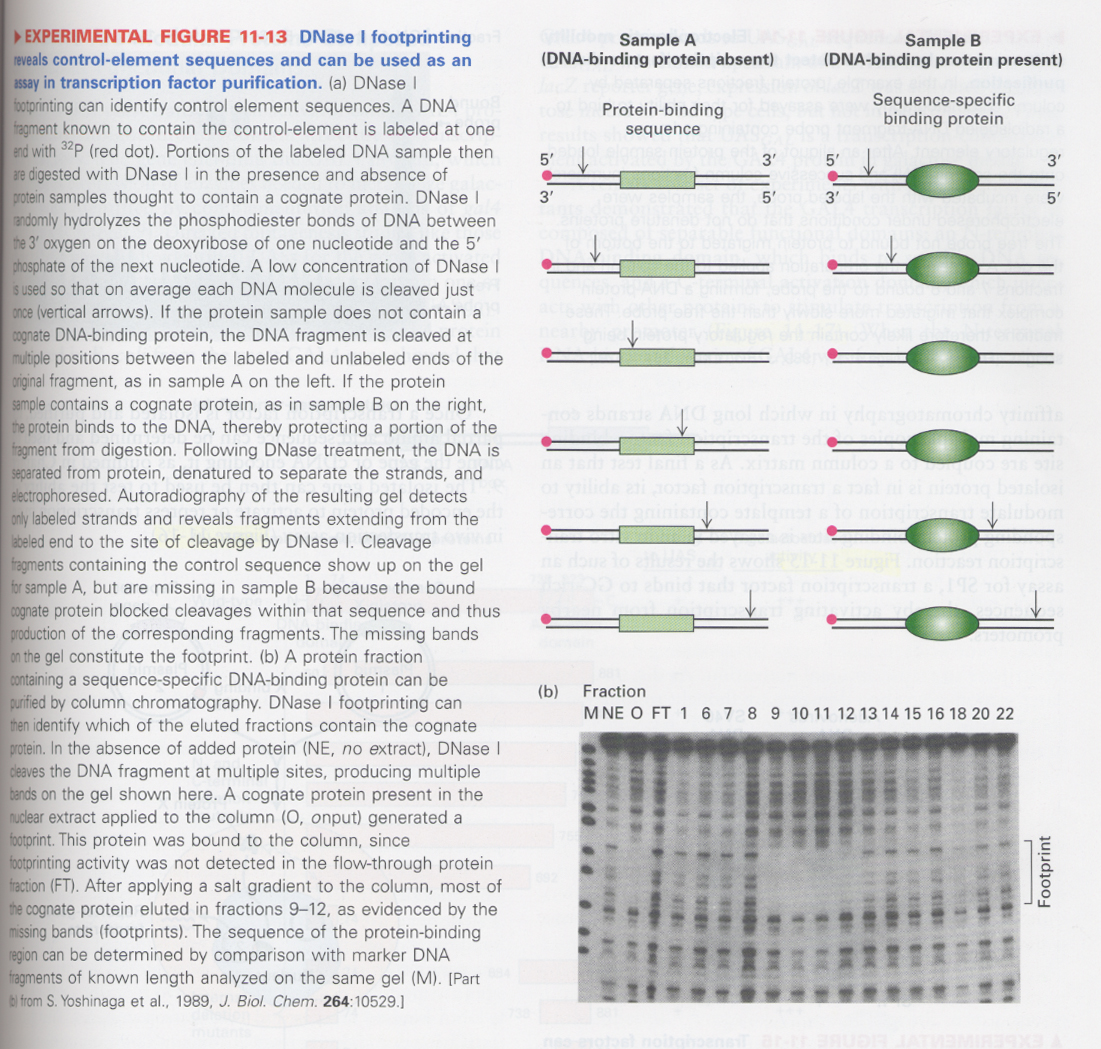
① DNase I footprinting
--> to see the protein binding sequence or for purification of DNA-binding proteins
--> NE ( absence of proteins), O (presence of proteins), FT (flow-through fraction)
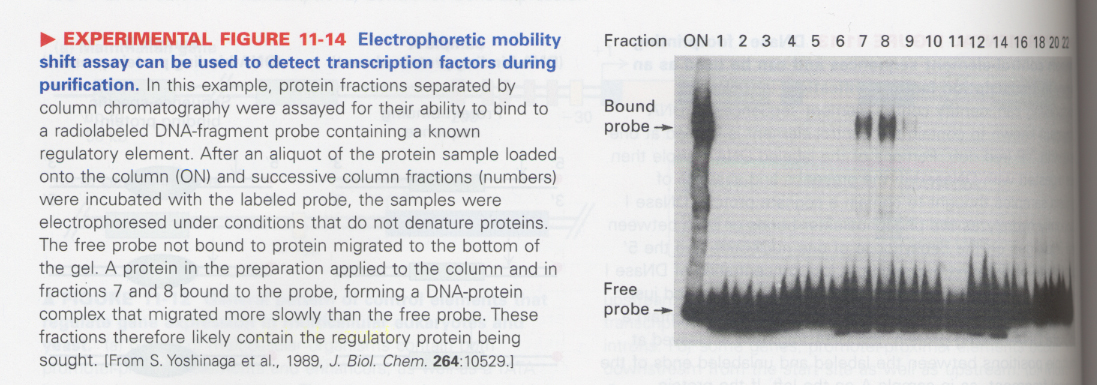
② electrophoretic mobility shift assay (EMSA)
--> for more quantitative analysis of DNA-binding proteins
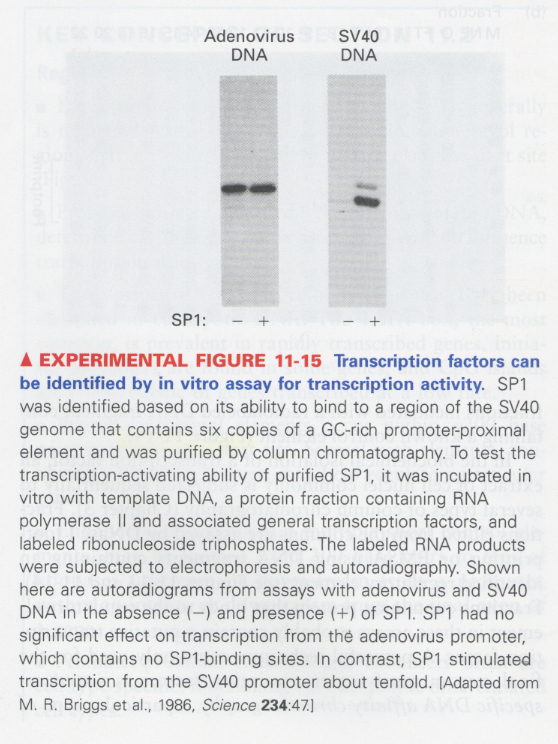
(8) in vitro transcription
--> to confirm whether the purified transcription factors have a transcriptional activity
--> adenovirus DNA does not have a SP1 binding site
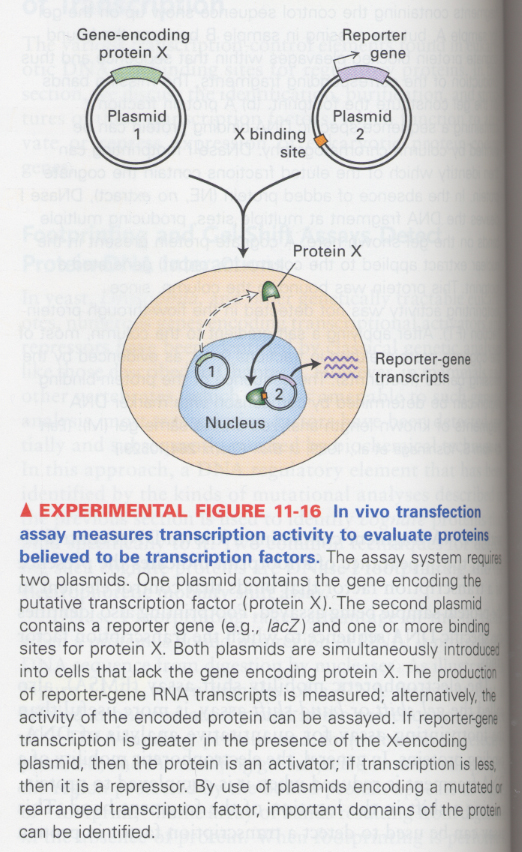
(9) in vivo transfection assay
--> to check whether the cloned gene of purified transcription factors is really able to transcribe
--> X is a cloned gene
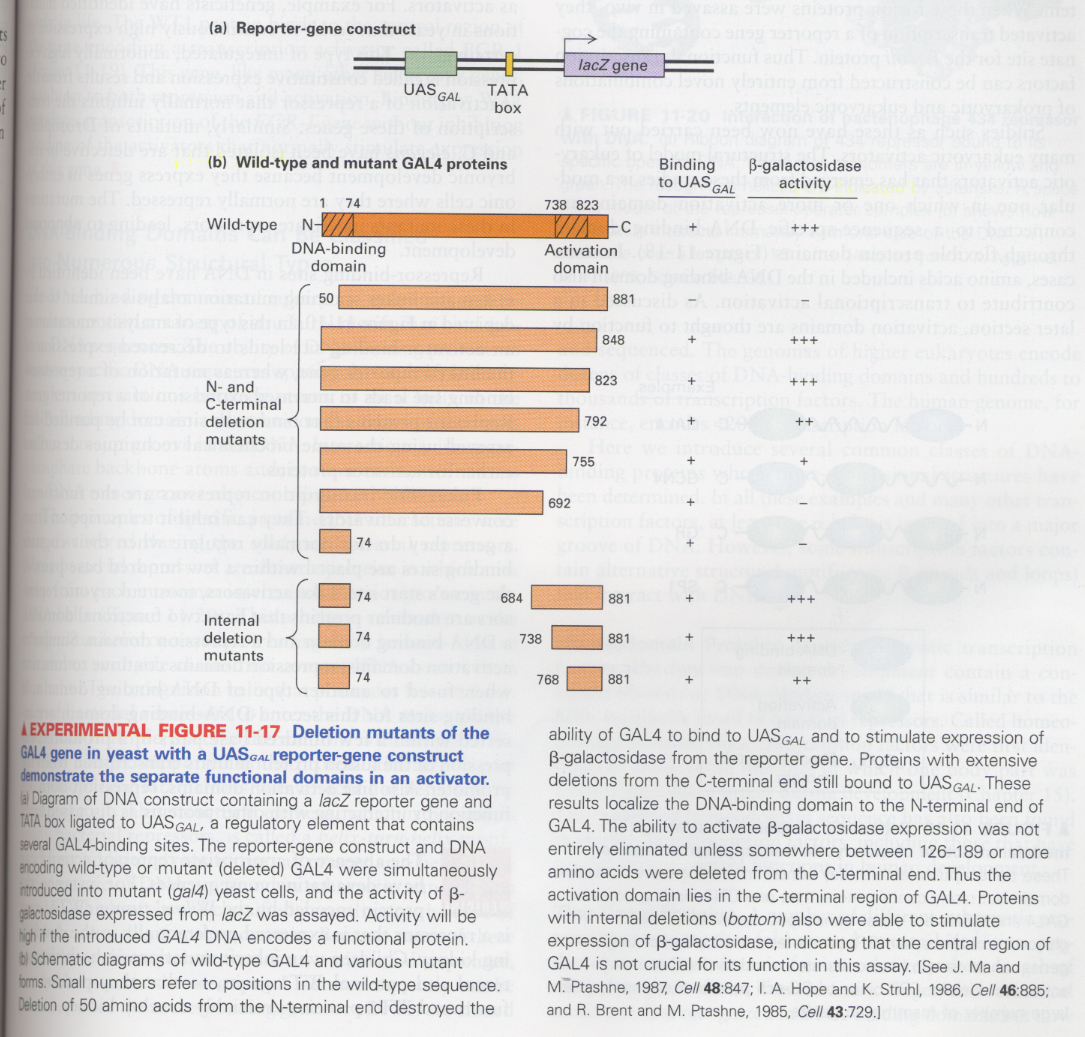
(10) Detection of functional domains in transcription factors (ex; GAL4) such as activator
--> from this experiment, two functional domains of GAL4 are discovered
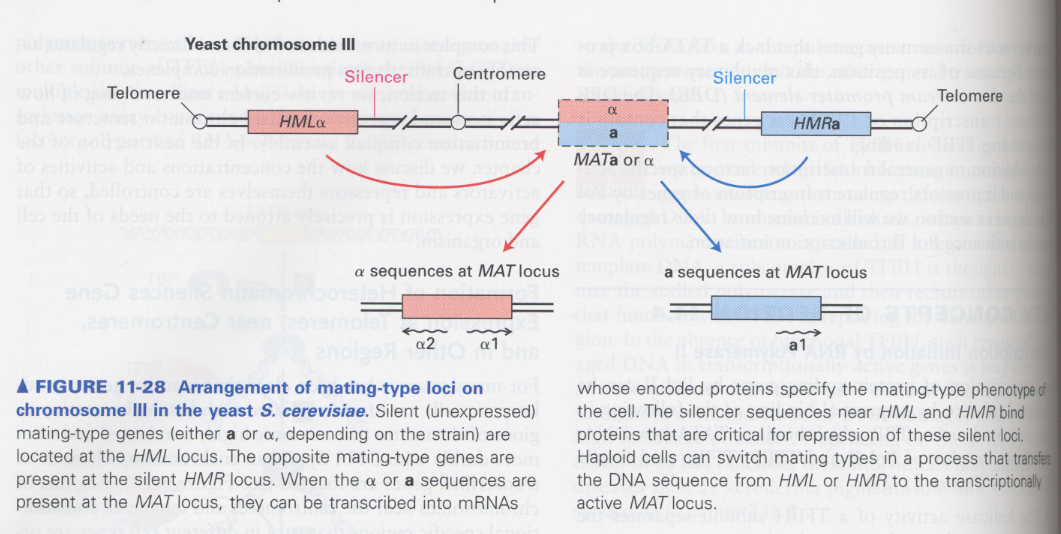
(11) mechanism of gene activation through exchage of chromosomal conformation
--> in yeast mating type; HMLα or HMRa with heterochromatin structure move into MAT with euchromatin
--> α and a type is determined
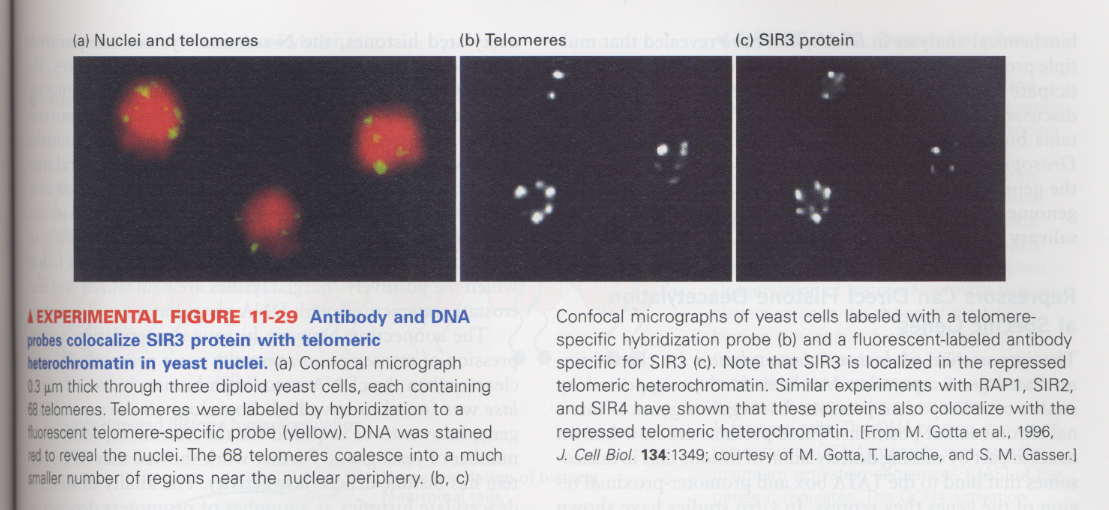
** repressor proteins involved in silencing; RAP1 and SIR proteins
--> colocalization of telomeres with SIR3
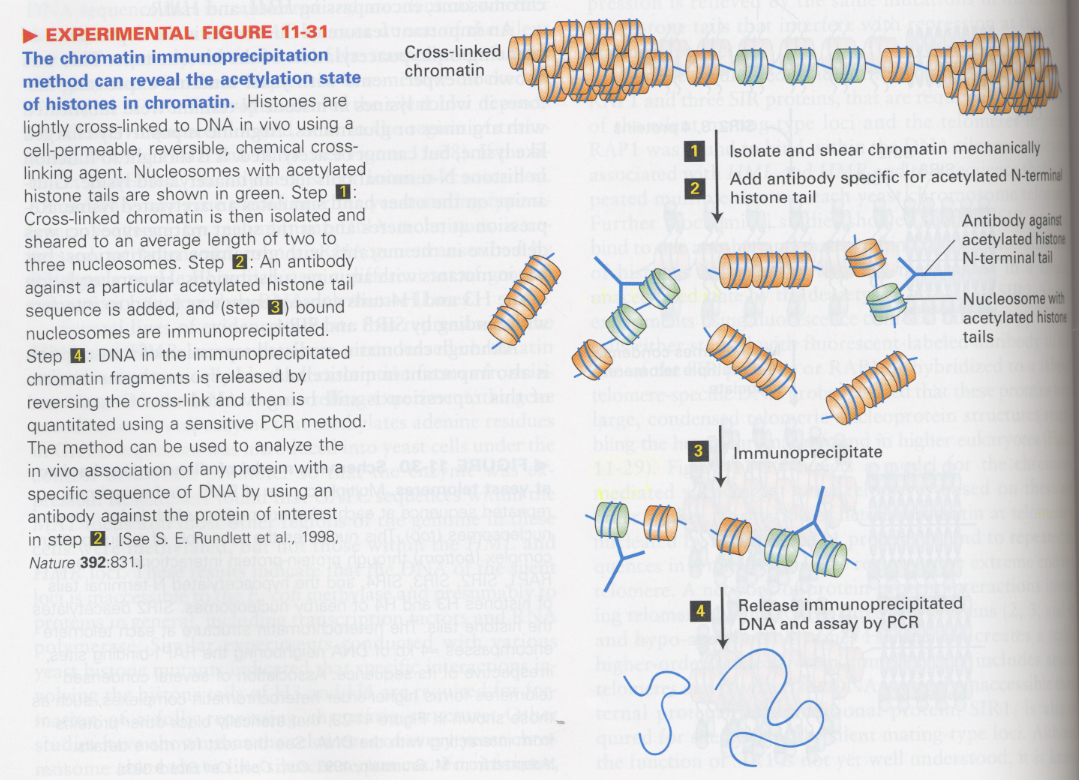
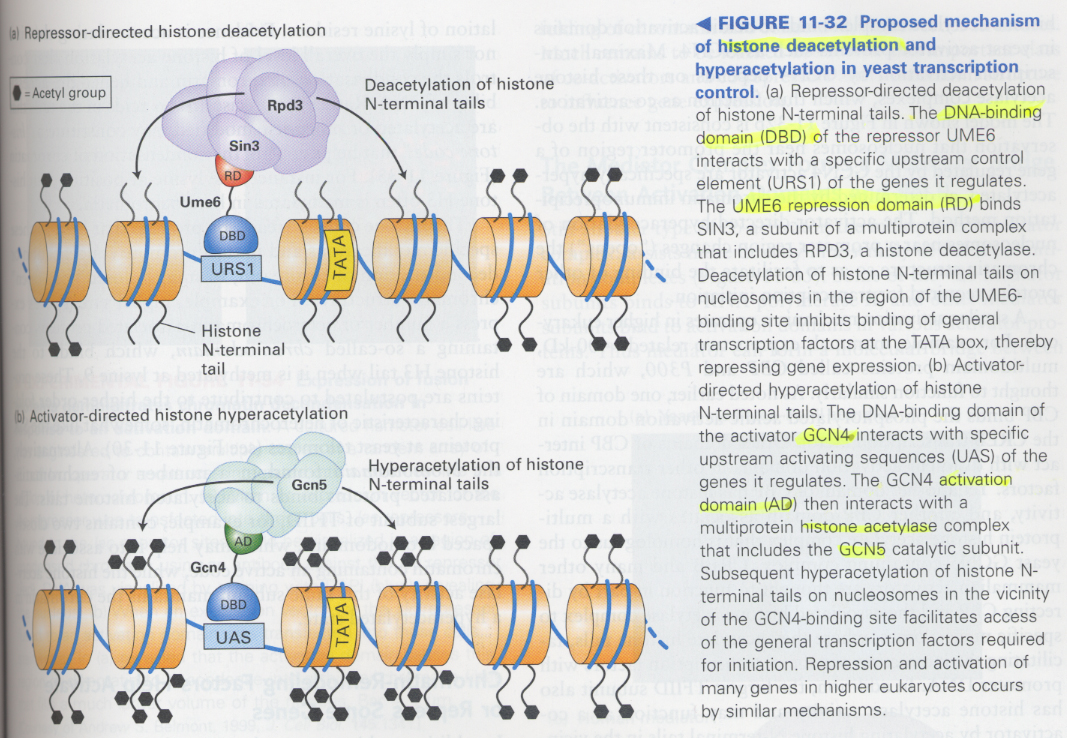
(12) Histone deacetylation
--> heterochromatin, untranscribed genes
** chromatin immunoprecipitation; to see the pattern of chromosomes
acetylation sites; N-terminal lysine residues of histones
** mechanism of histone deacetylation and acetylation
① histone deacetylation; inactivation, Ume6, Sin3, RPD3 (deacetylase)
② histone acetylation; activation, Gcn4, Gcn5 (acetylase)
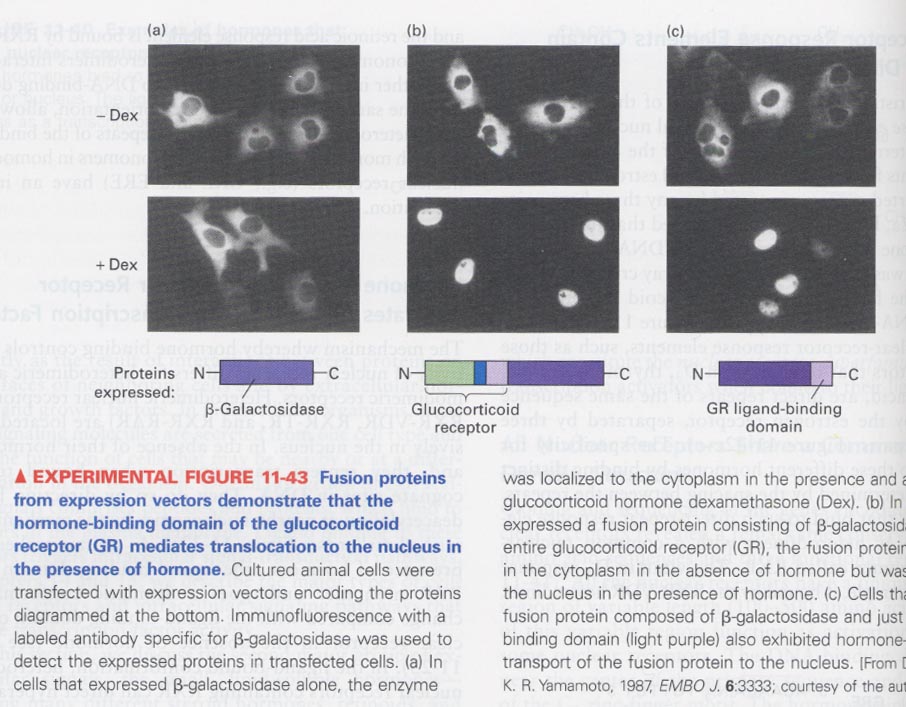
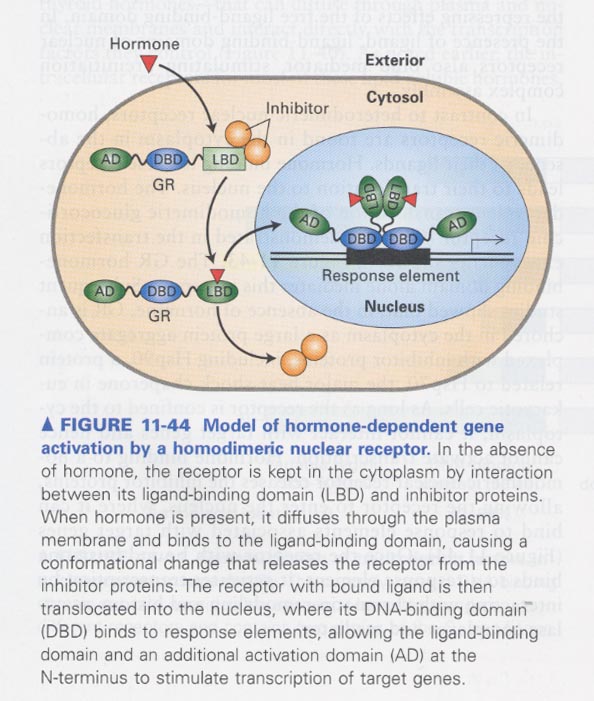
(13) translocation of homodimeric glucocorticoid receptor (GR)
--> from cytosol to nucleus in the presence of dexamethasone
** hormone-dependent gene activation
12장; Post-transcriptional control and nuclear transport
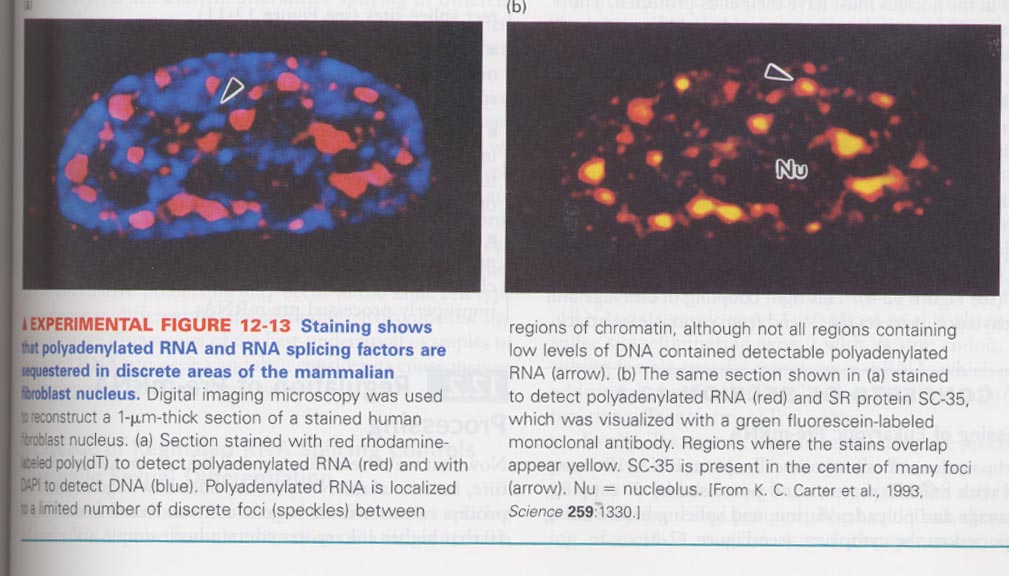
(1) RNA splicing
--> DNA:mRNA hybridization reveals the presence of introns
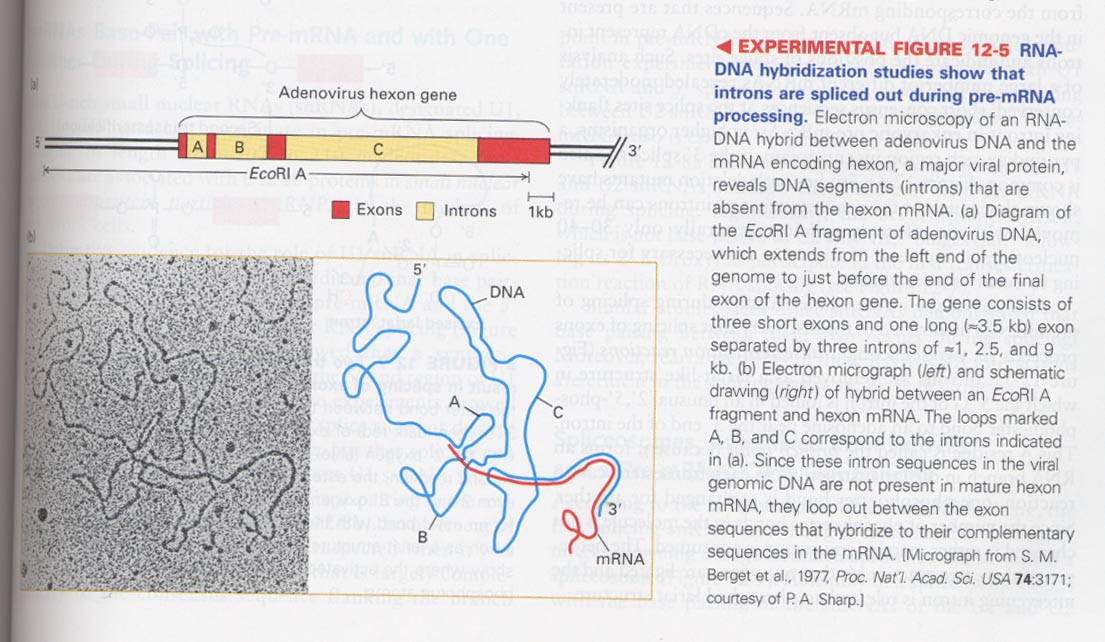
--> splicing sites; poly (T) (red), DAPI (blue), SR protein (green)
--> splicing is occurred at discreted areas of nucleus
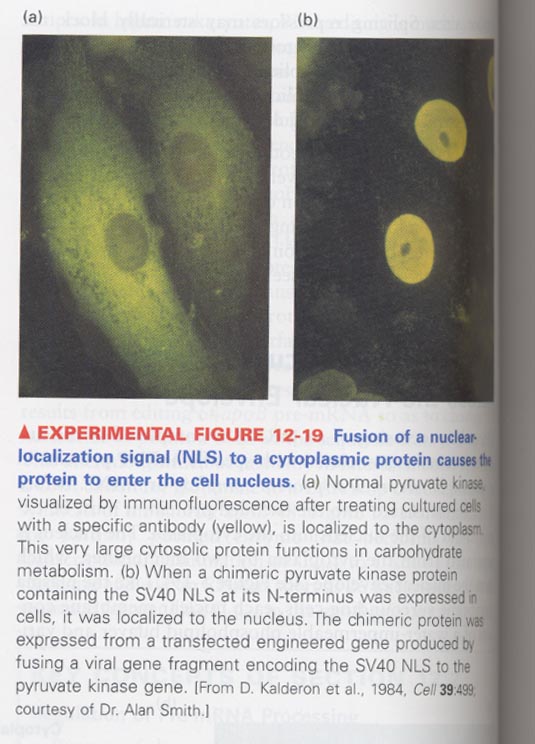
(2) Role of nuclear localization signal sequence (NLS)
--> NLS origin; come from T antigen of SV40 (wt type is present at a nucleus, mt type is at cytoplasm)
--> basic residues are rich (Pro-Lys-Lys-Lys-Arg-Lys-Val)
** cytosolic pyruvate kinase + NLS of SV40 T antigen --> movement into nucleus
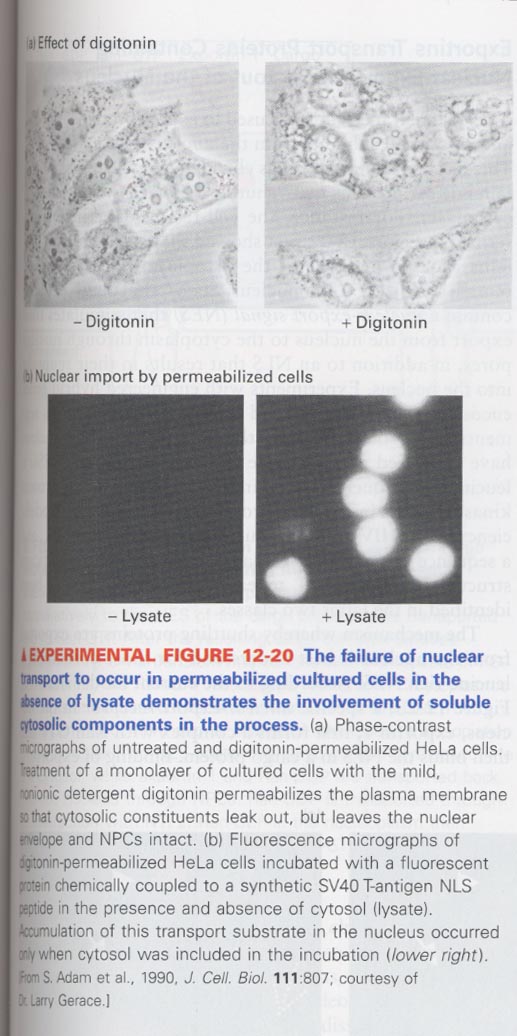
(3) Requirement of cytoplasmic proteins for nuclear transport
--> cytoplamic proteins; Ran, NTF2 (nuclear transport factor 2), importinα and β
--> experiment; digitonin treatment causes a permeabilization of plasma membrane, bu intact in nuclear envelopes
** fluorescent protein + NLS
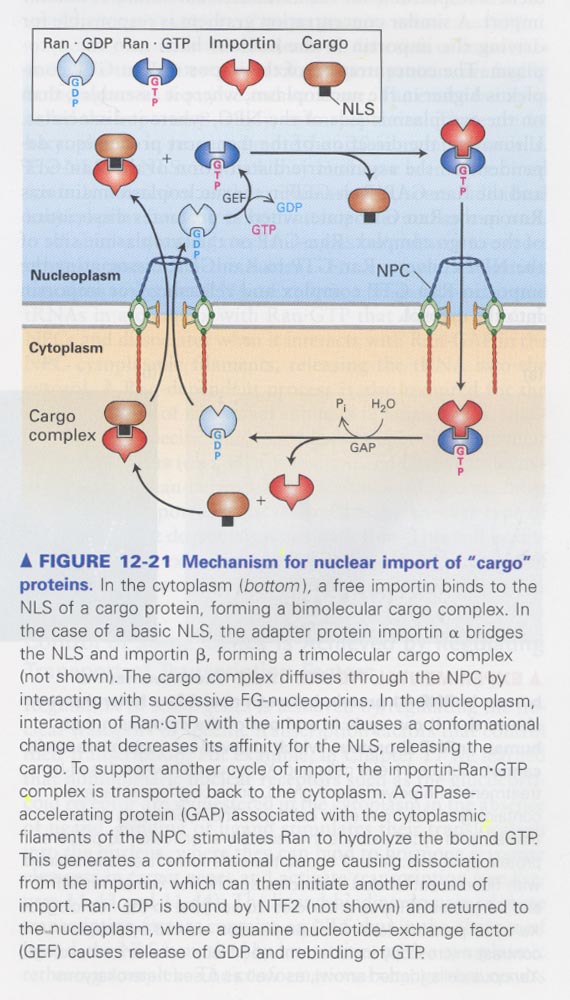
** nuclear import model
(3) nuclear export
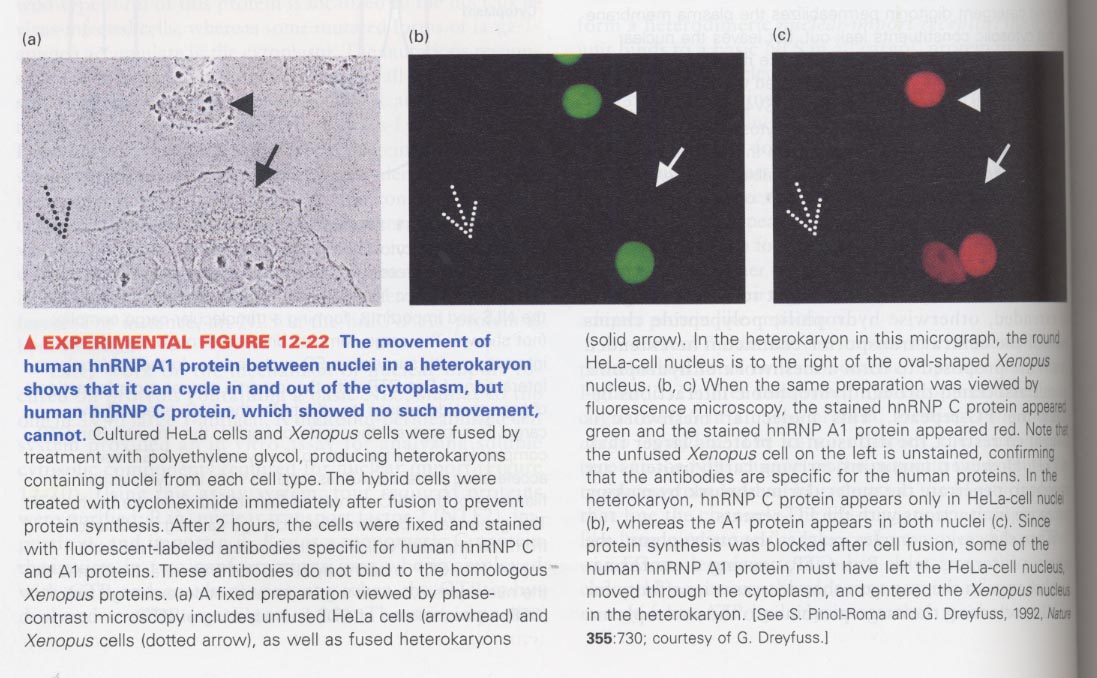
--> cell fusion experiment; HeLa cells + Xenopus cells
green (for human hnRNP C), red (for human hnRNP A1)
--> NES (nuclear export signal); ① leucine-rich sequence ② a 38-residue sequence ③ a sequence in hnRNP K
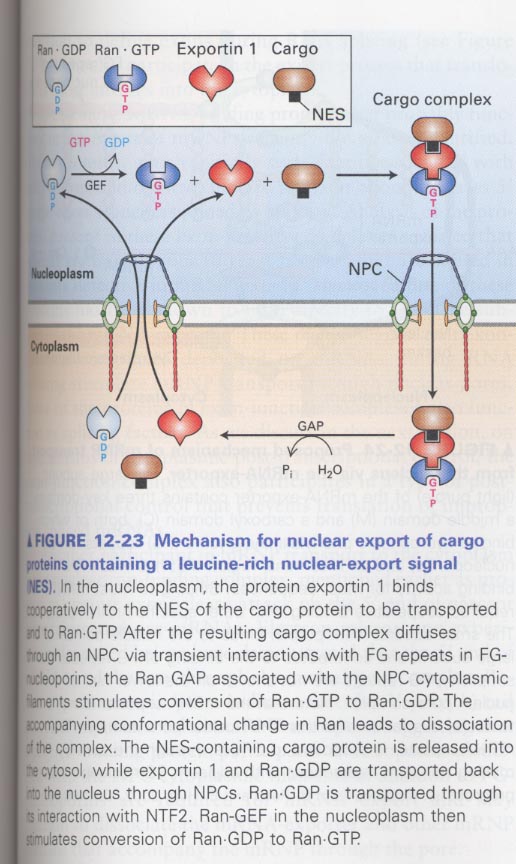
** nuclear export model; exportin1 + Ran.GTP --> + NES of cargo proteins
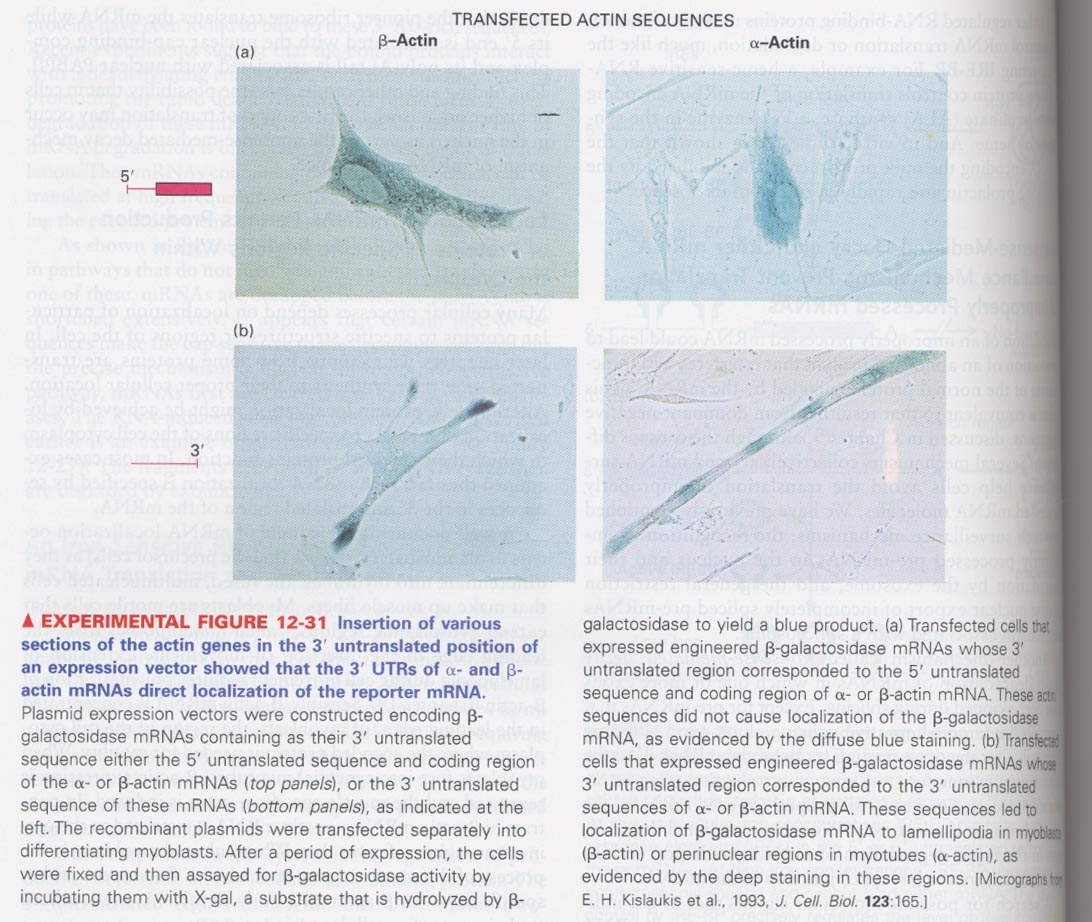
(4) Role of 3'UTR of mRNA in the protein targeting
--> β actin is localized in the leading edges, α actin is localized into the perinuclear regions of myotubes
--> β-galactosidase (5') + (3') actin regions in pannels
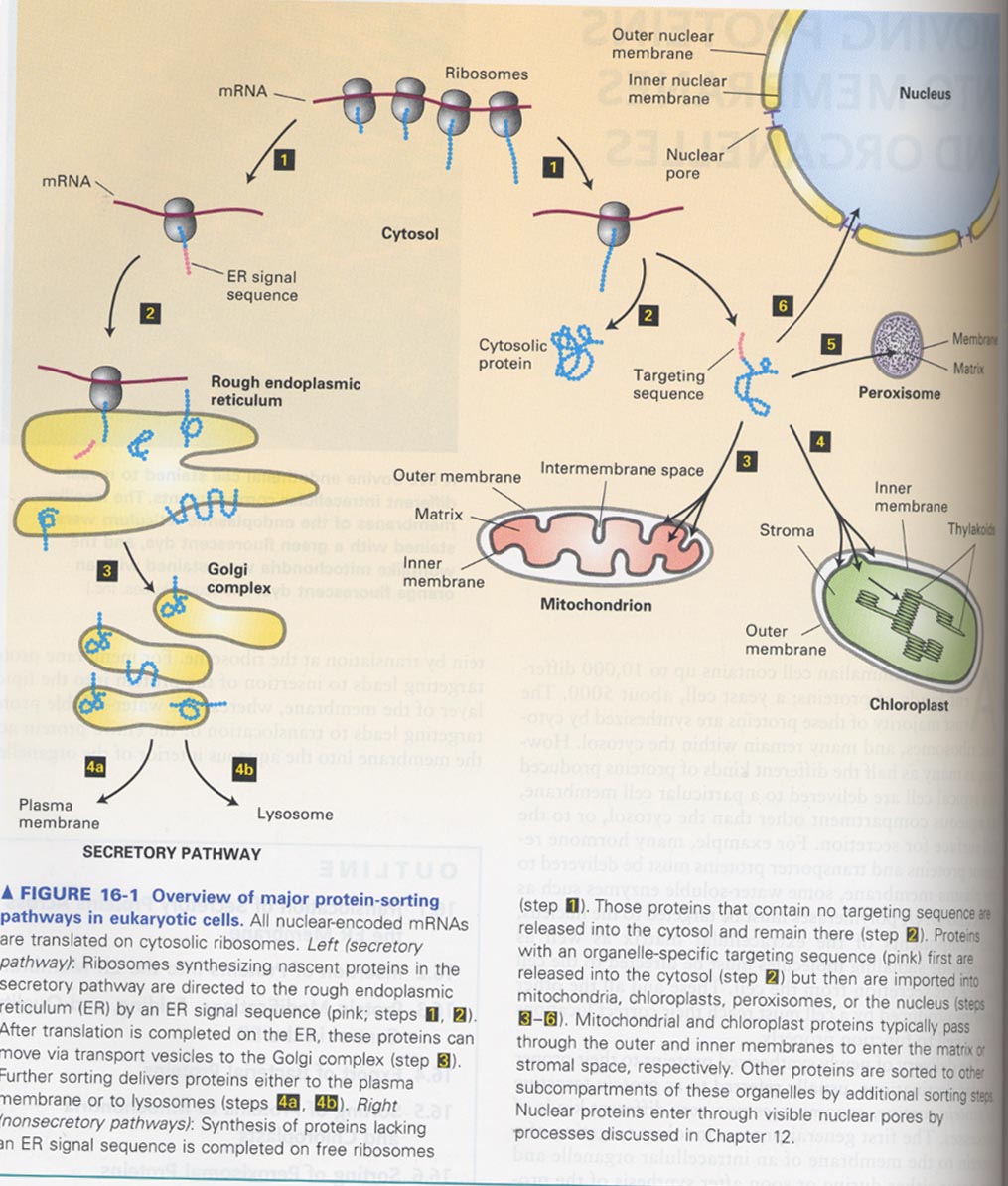
16장; Moving proteins into membranes and organelles
(1) protein sorting pathways
--> secretory pathway, nonsecretory pathway
(2) secretory protein localization in the lumen of ER
--> pulse-labeling experiment; generation of rough microsomes
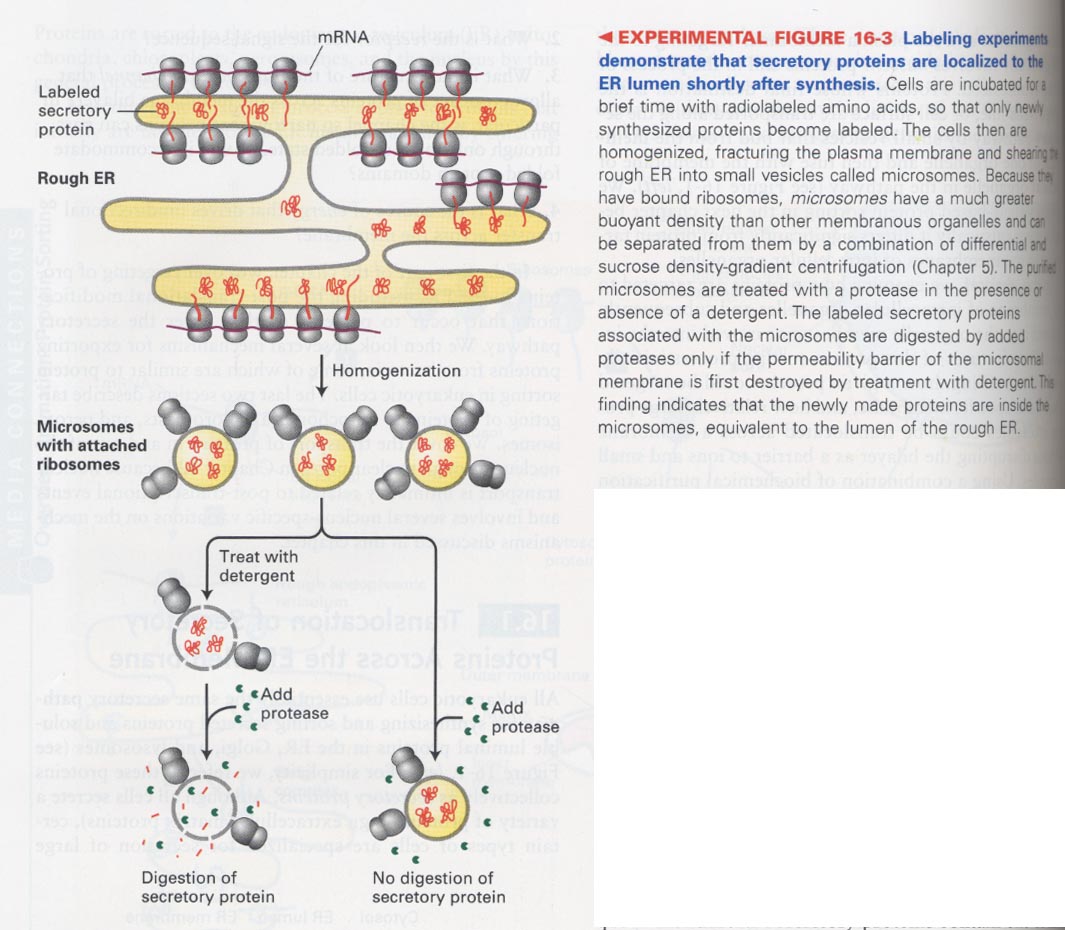
(3) hydrophobic N-terminal sequence (ER signal sequence) is associated with microsomes
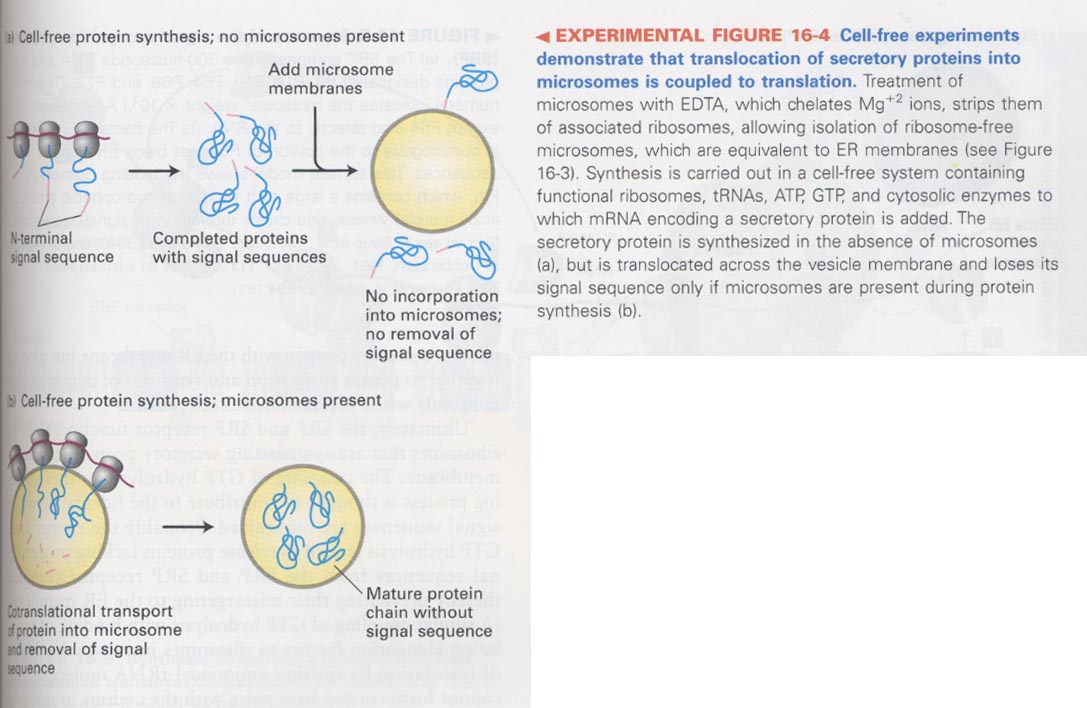
--> cell-free experiments
--> ER signal sequence; 1-2 positive charged Aas + 6-12 hydrophobic core sequence
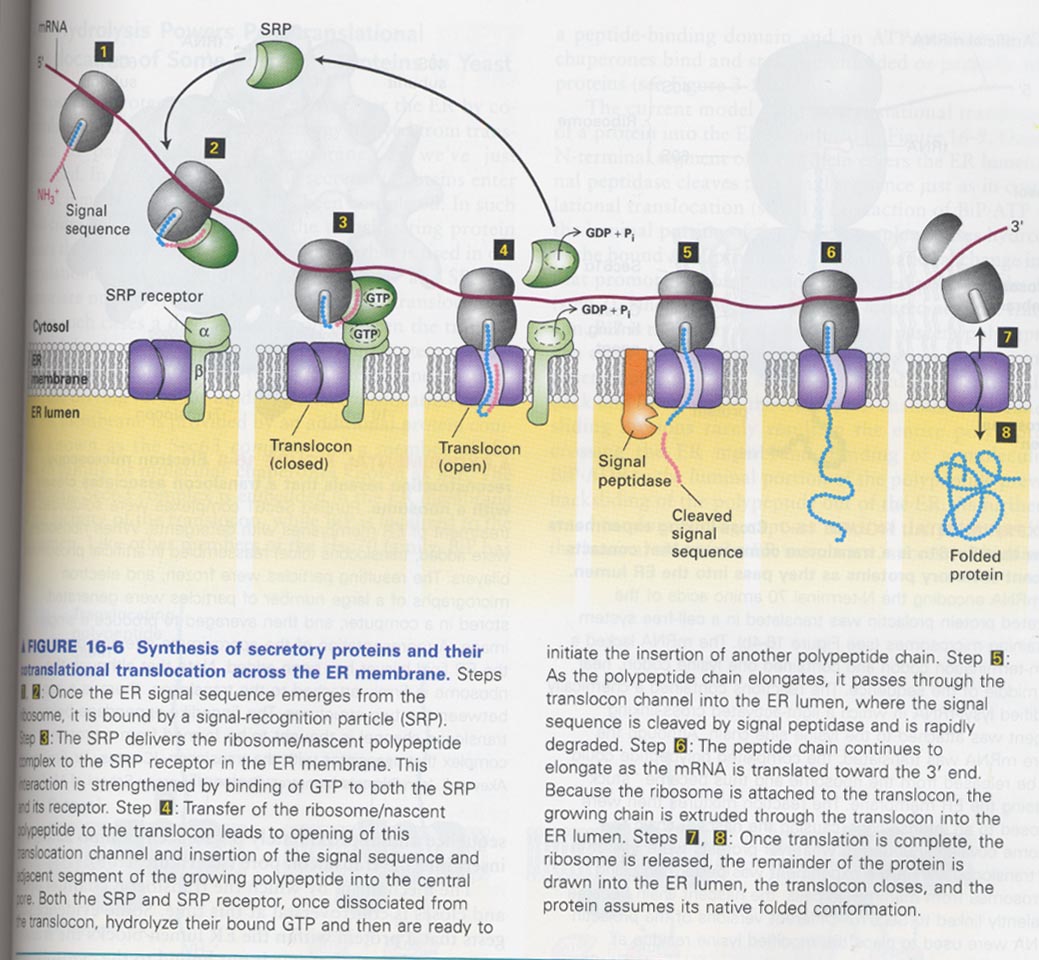
(4) ER translocation
** Model for ER translocation
--> SRP (signal recognition particle)
--> translocon; 3 different proteins (Sec61α, Sec61β, Sec61γ)
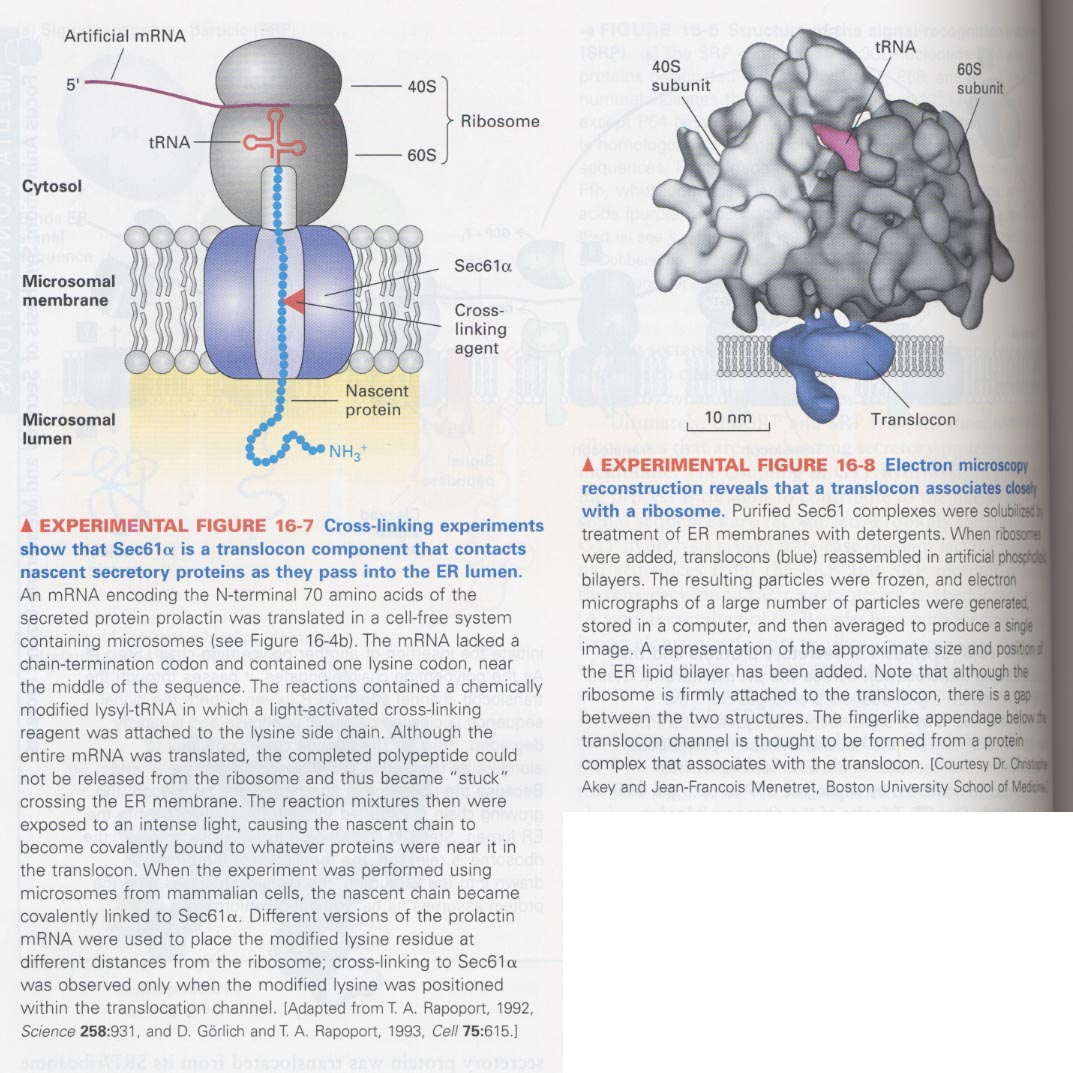
--> translocated proteins are contacted with Sec61α among Sec61 complex
; use of modified Lys-tRNA binding to the light-activated cross-linking reagent
(5) Resolution of the topology of membrane proteins from their sequences
--> positive; hydrophobic portion, negative; polar portion
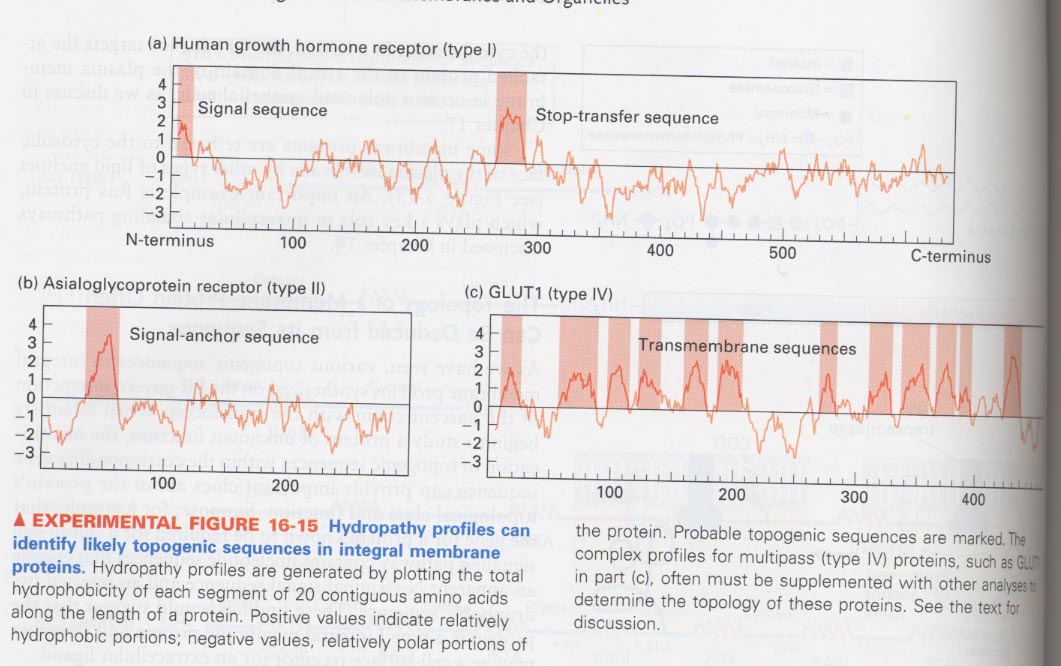
(6) mitochondrial matrix-targeting sequences
--> matrix-targeting sequence; at the N-terminal, 20-25 Aas,
hydrophobic Aas + basic Aas + hydroxylated Aas (Ser, Thr)
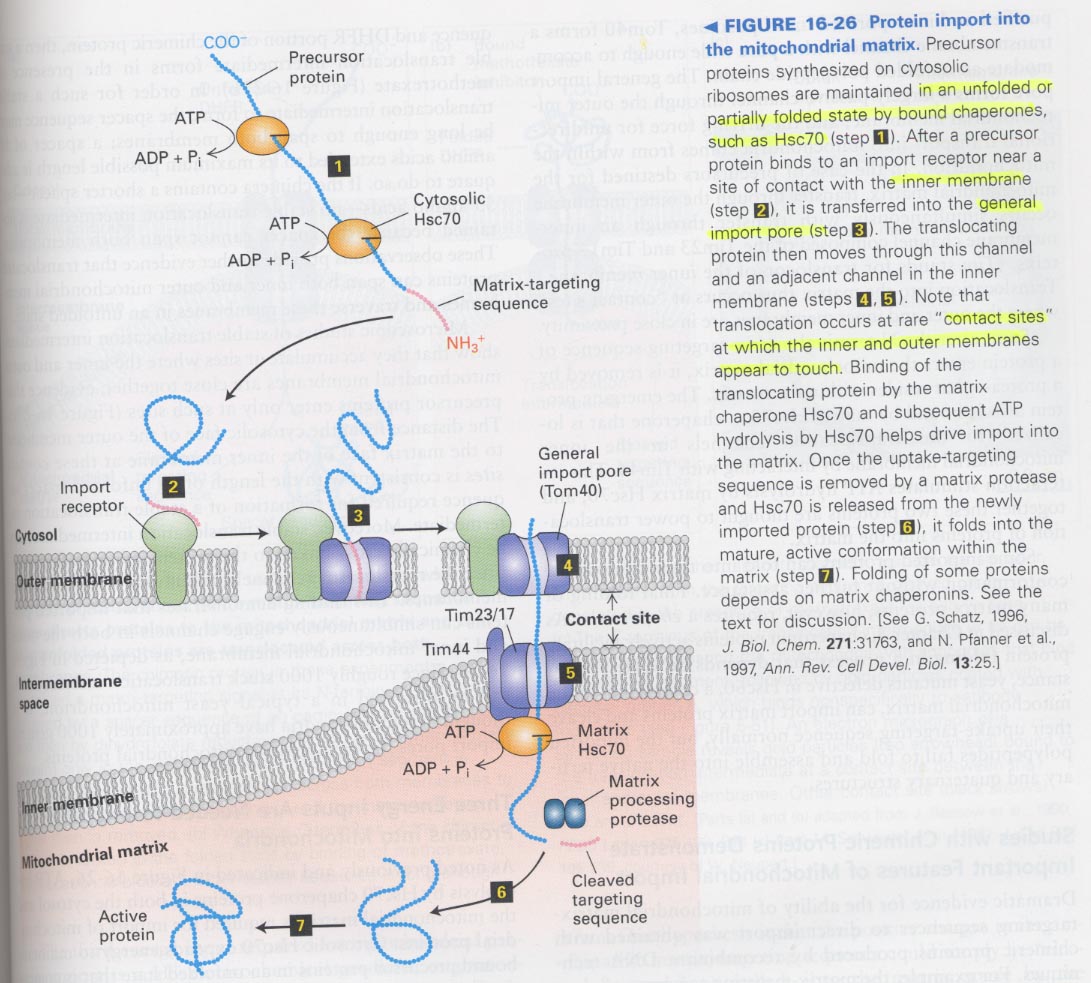
** Model for mitochondrial protein targeting
--> cytosolic HSP70; for unfolding, import receptor, import pore (Tom40, Tim23/17, Tim44)
matrix HSP70; ATP hydrolysis cause to pull out the translocated protein
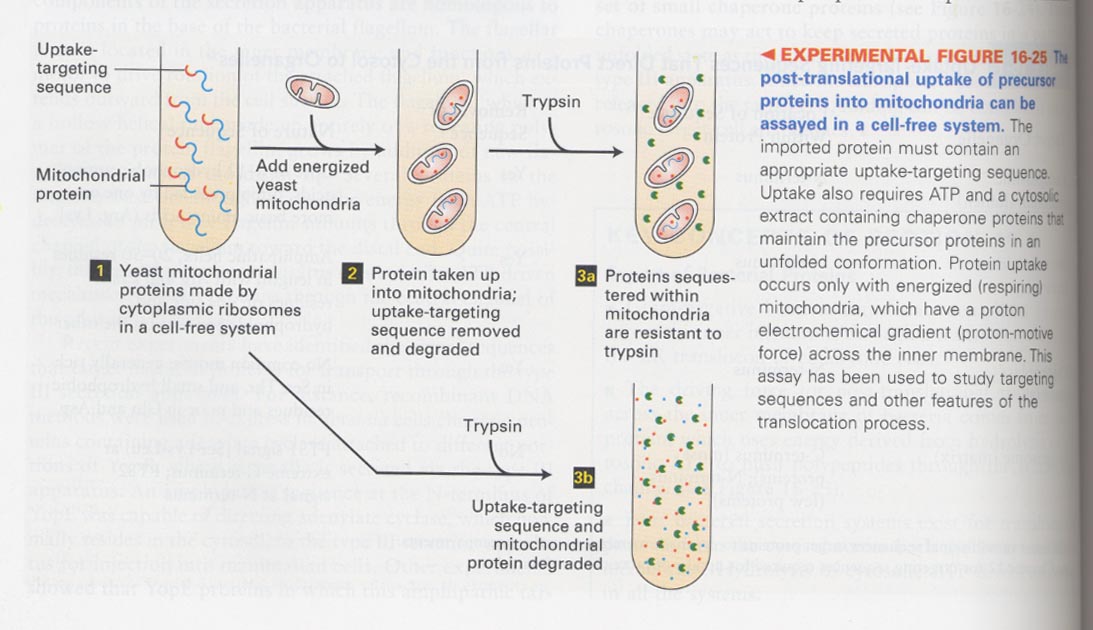
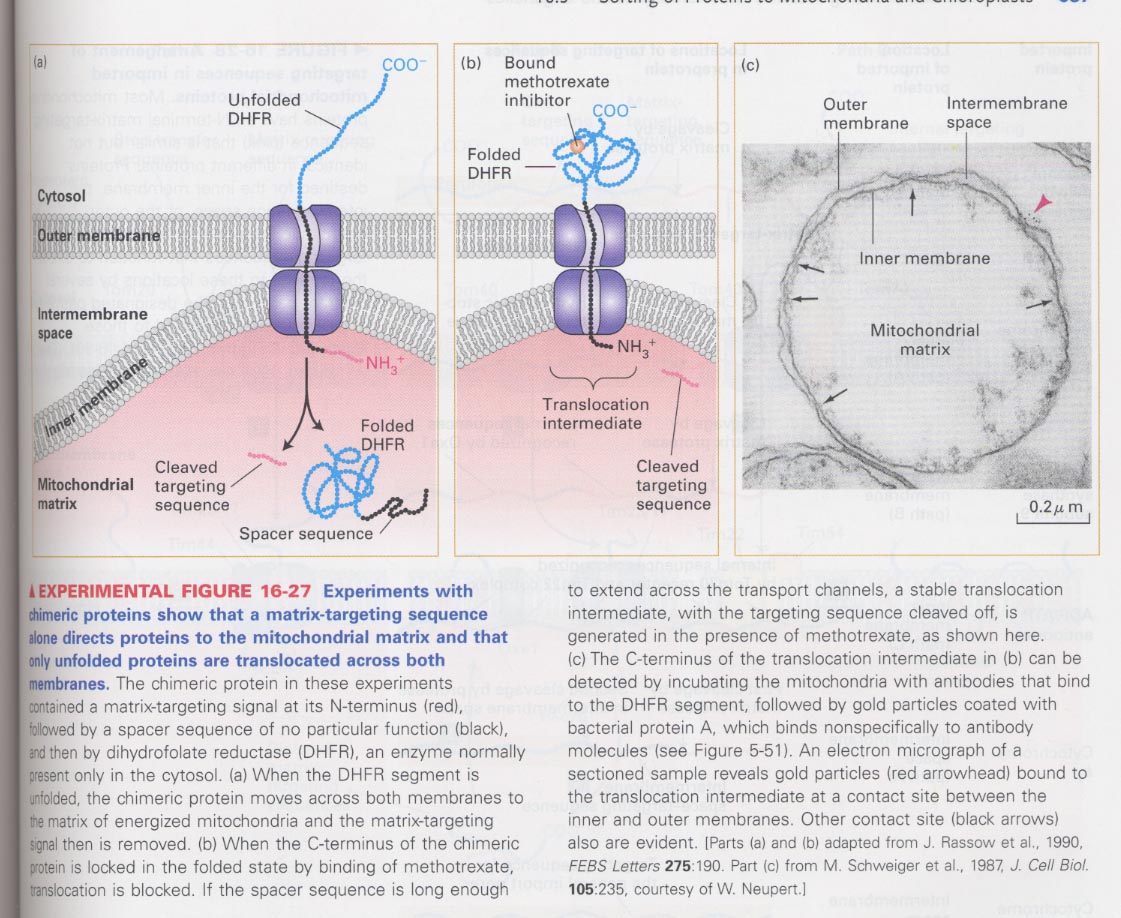
(7) Role of mitochondrial matrix-targeting sequences in the protein targeting
** cell-free translocation assay;
matrix-targeting sequences of alcohol dehydrogenase + a spacer sequence + dihydrofolate reductase (DHFR)
--> in the presence of chaperones; unfolding causes the translocation
in the presence of methotrexate; folding causes the non-translocation
--> (c) ; presence of (b) using antibody against DHFR
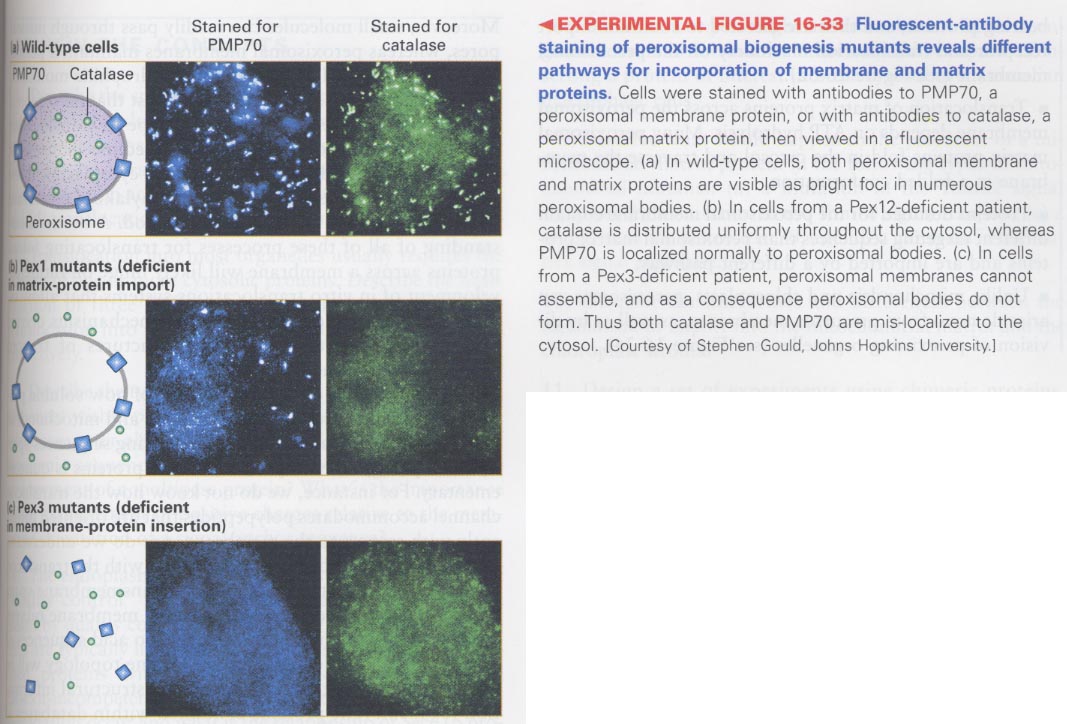
(8) peroxisomal proteins
--> Zellweger syndrome; defect in transport of peroxisomal matrix proteins
--> for peroxisomal matrix proteins; Pex10, Pex12, Pex2
--> for peroxisomal membrane proteins; Pex3, Pex16
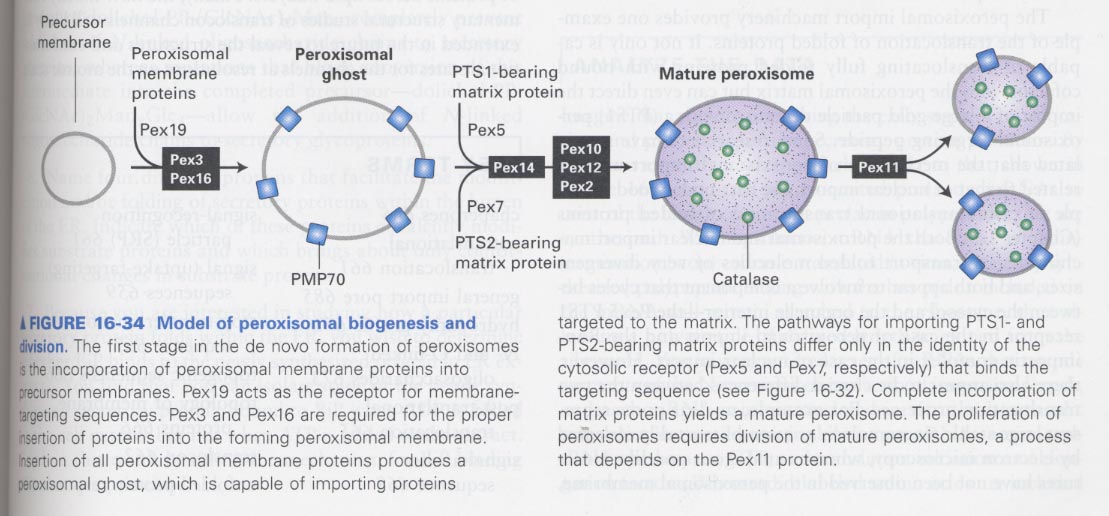
** peroxisomal biogenesis
--> Pex19 act as a receptor
--> PTS1 (peroxisome targeting sequence) sequence; C-terminal, PTS2 sequence; N-terminal
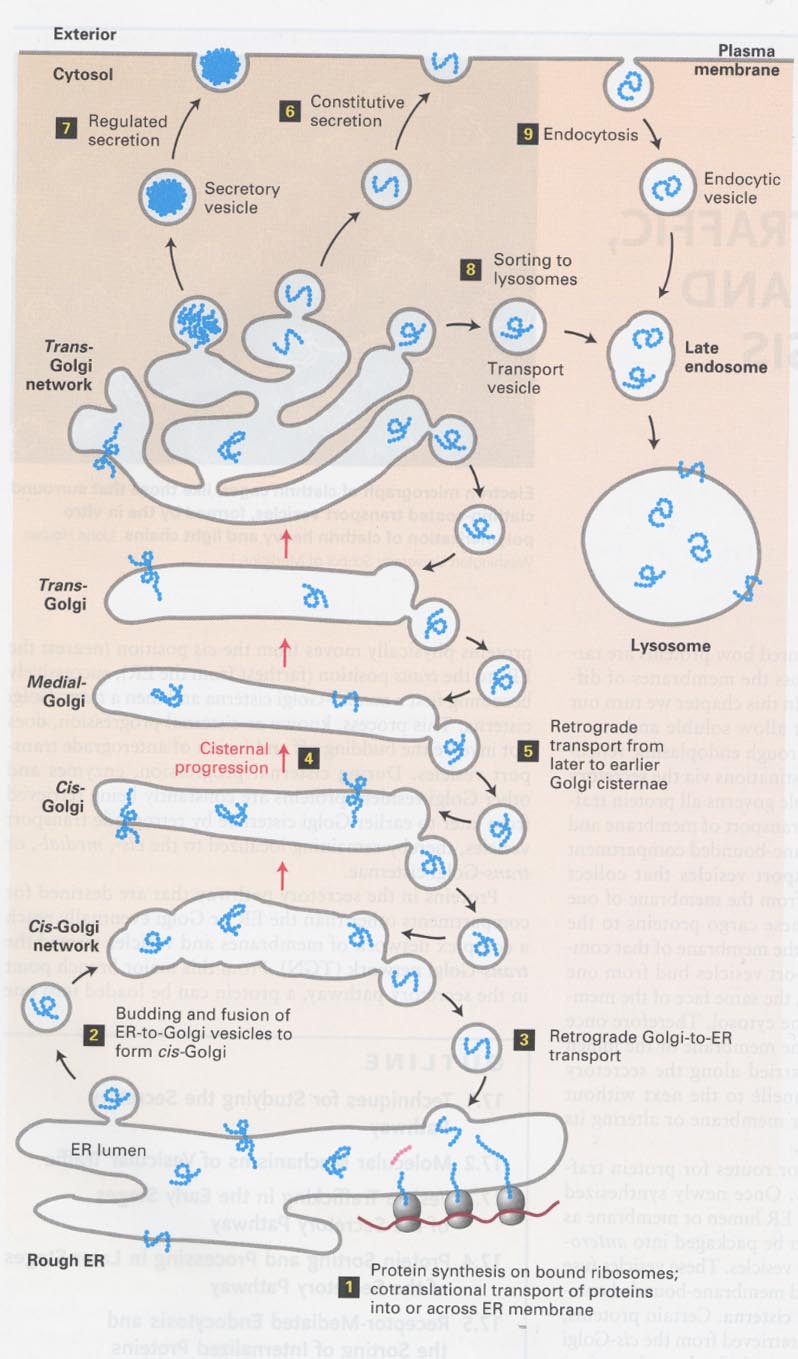
17장; Vesicular traffic, secretion, endocytosis
(1) secretory and endocytic pathways of proteins
--> cisternal progression; a nonvesicular process
--> retrograde transport; ER-or Golgi-resident proteins
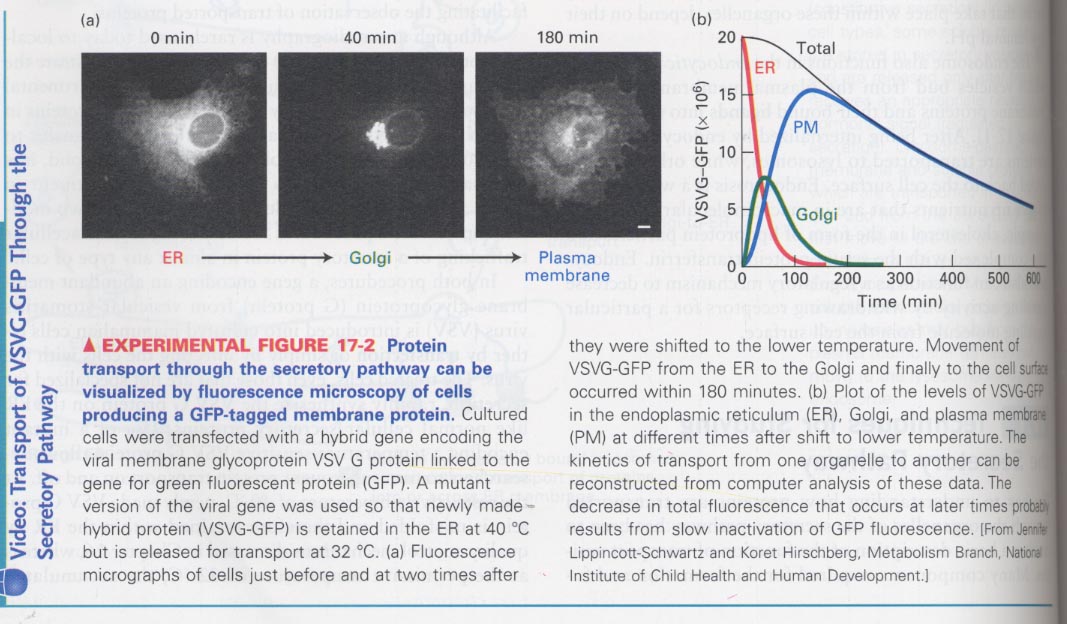
(2) Use of GFP fusion proteins that is sensitive to Temp.
--> for observation of secretory proteins
--> ex) glycoprotein of vesicular stomatitis virus (VSVG)
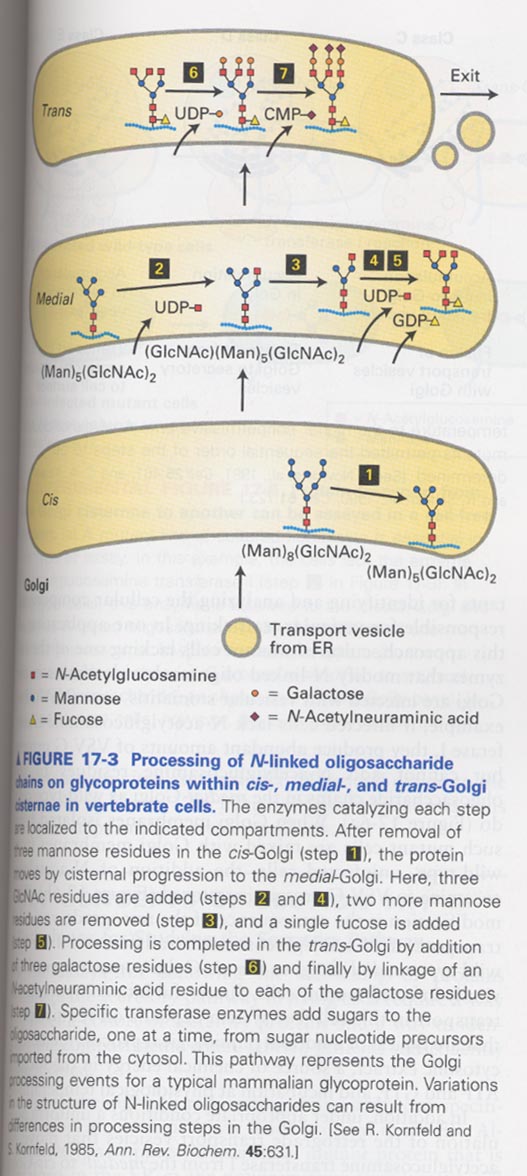
(3) Compartment-specific oligosaccharide modification
--> (Man)8(GlcNac)2
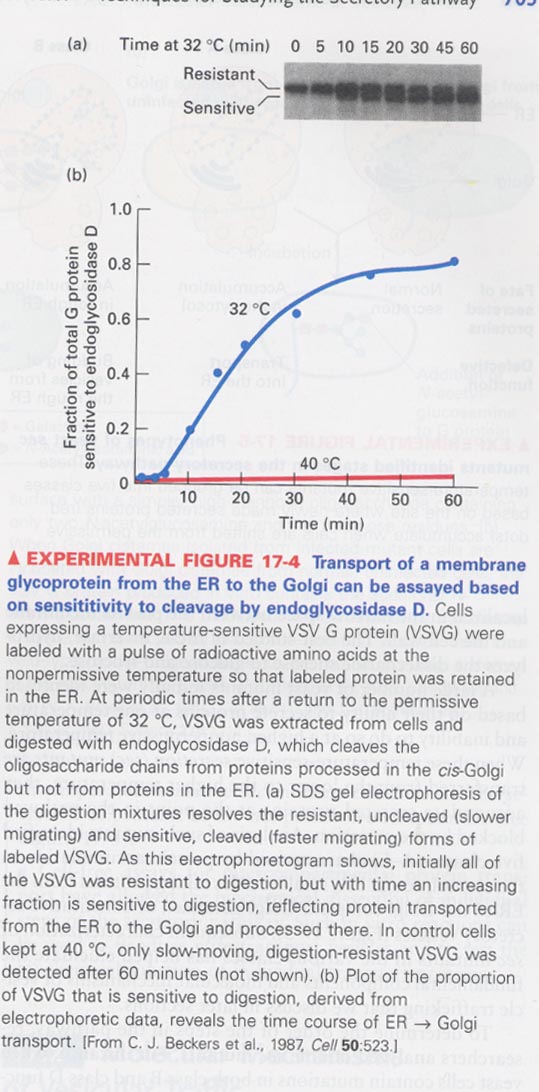
--> Assay for a glycoprotein transport from ER to cis-Golgi
--> pulse-labeling experiment; label in the nonpermissive Tem. (40oC) and treated with endoglycosidase D (cis-Golgi
specific)
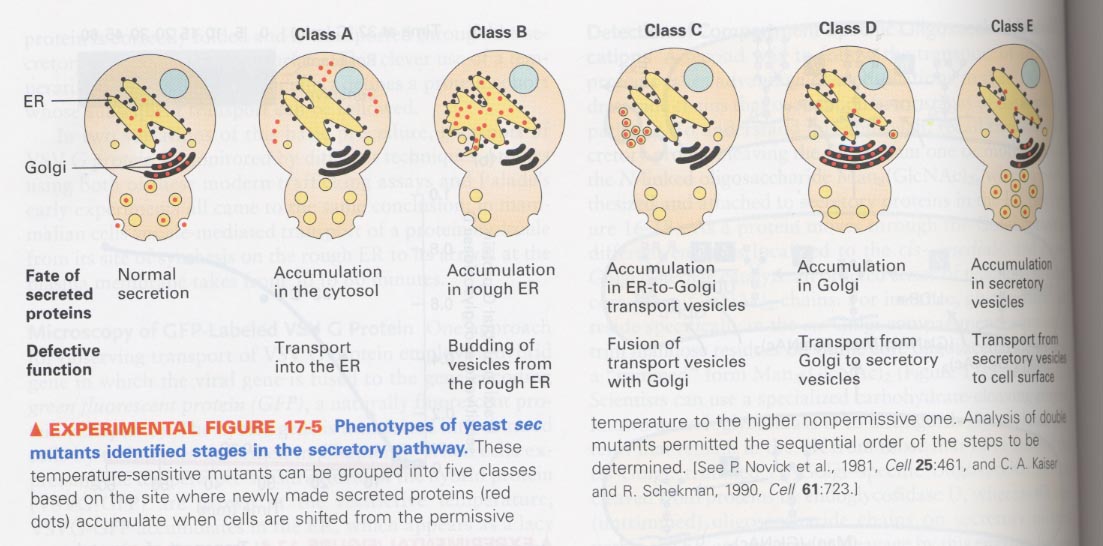
(4) Sec mutants reveal the stages of secretory pathway
--> 5 kinds of sec yeast mutants
--> characterization of accumulated proteins when the mutant is shifted from permissive to nonpermissive Temp.
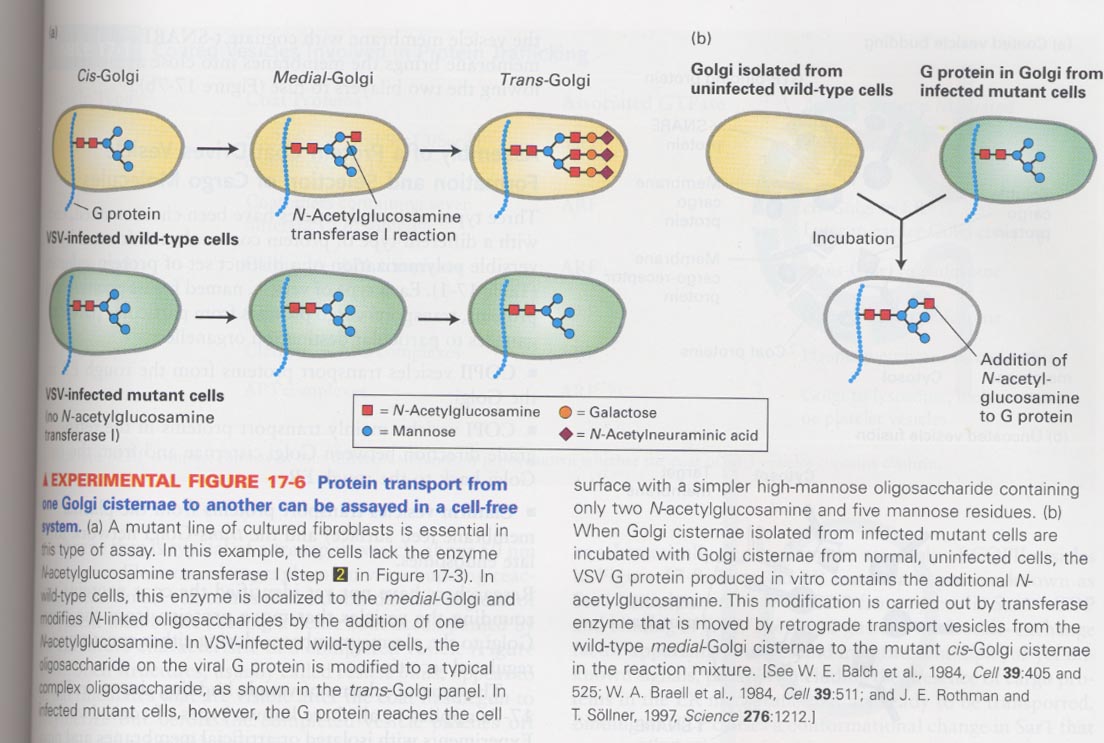
(5) Protein transport through Golgi compartments
--> cell-free transport assays; fibroblast lacking N-acetylglucosamine transferase I + VSV G protein
(6) Basic mechanism underlying vesicle budding and fusion
--> (a) budding, (b) fusion
--> in vitro budding reaction; polymerization of the coat proteins (dark regions)
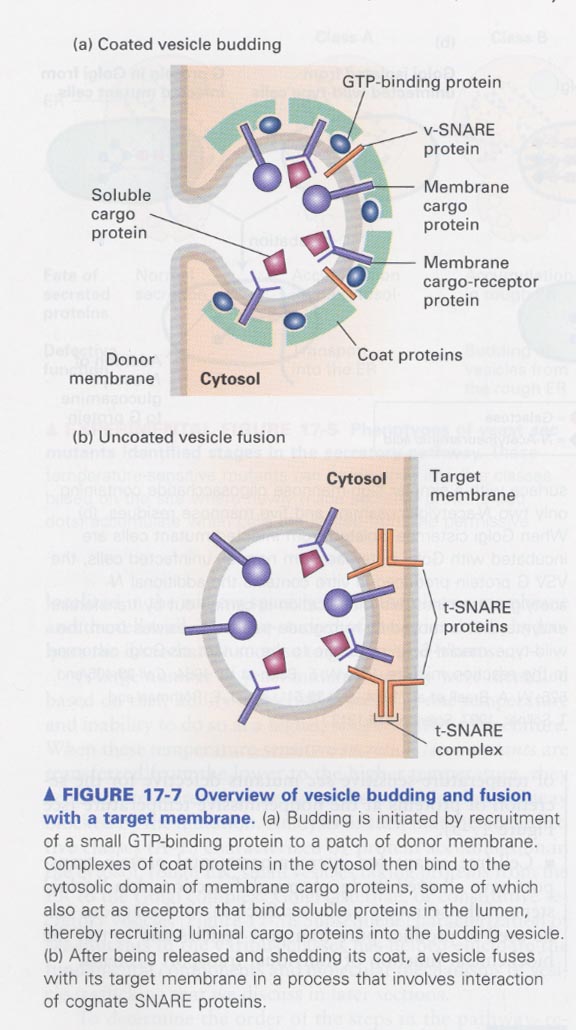

(7) Role of GTP-binding proteins in the assembly of vesicle coats
--> 3 kinds of vesicles; ① COPII; ER->Golgi ② COPI; between Golgi, cis-Golgi->ER
③ clathrin; membrane, trans-Golgi->endosome
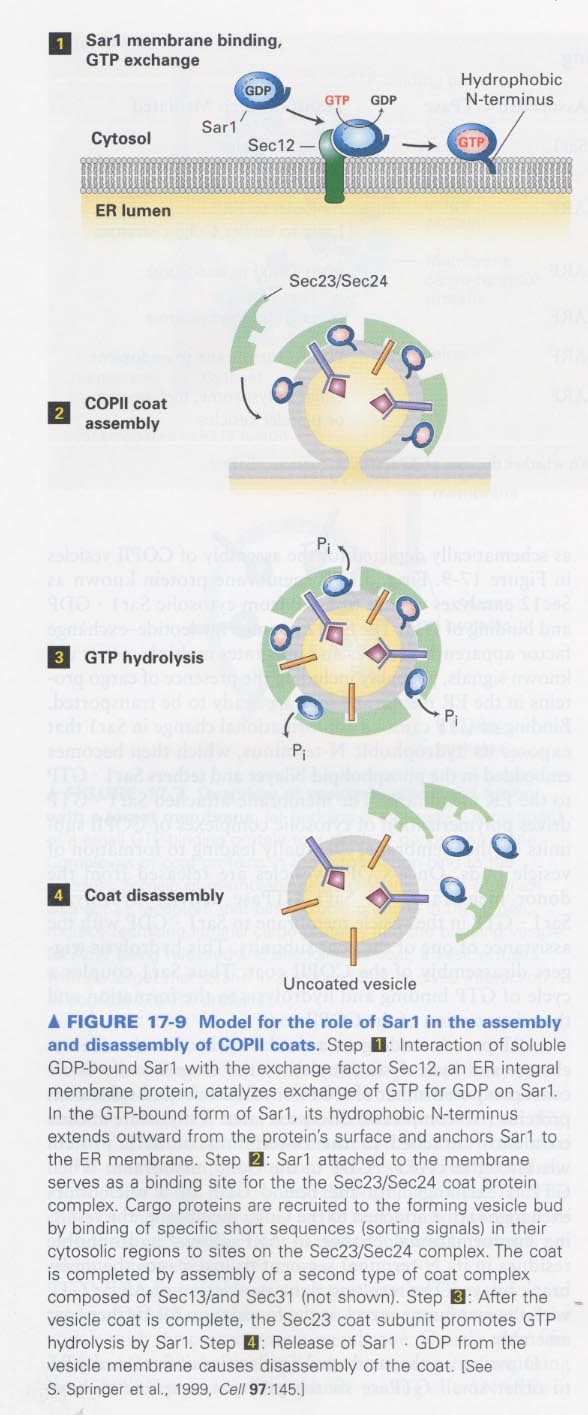
** Model for vesicle assembly and disassembly
(1) Sar1 (small GTP-binding protein; a regulatory function. Ras like) + Sec12 (exchange factor)
(2) Sec23/24 binding to GTP-Sar1
(3) Cargo protein binding to Sec23/24
(4) Sec13/31 binding --> completion of a coat complex
(5) GDP-Sar1 formation by Sec23
(6) Coat disassembly
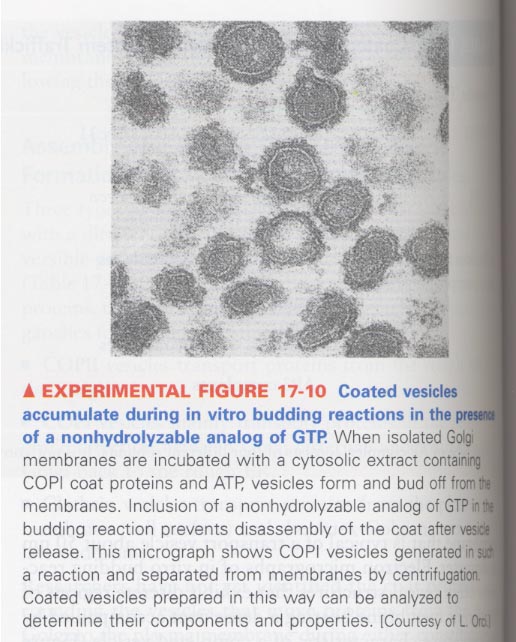
--> mutant version of Sar1 in the GTP hydolysis
; no disassembly of coat and unable to fuse with target membranes
ex) addition of a nonhydrolyzable GTP analog
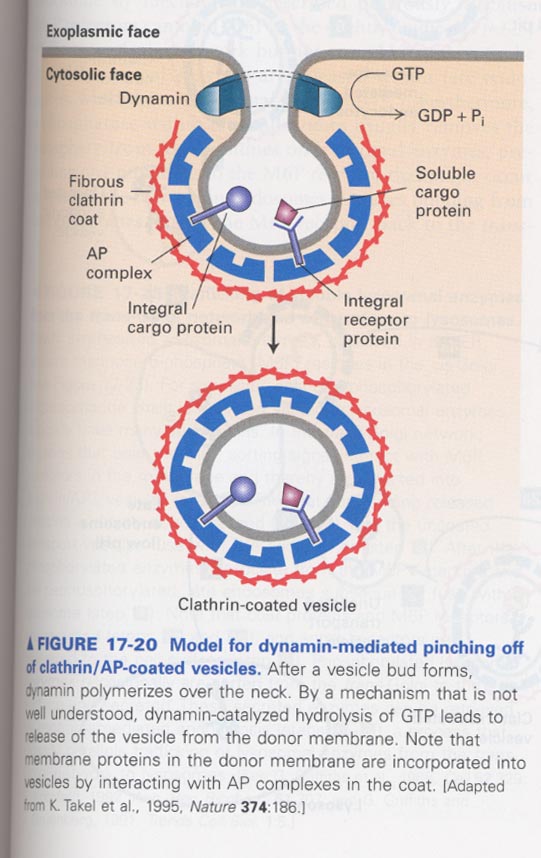
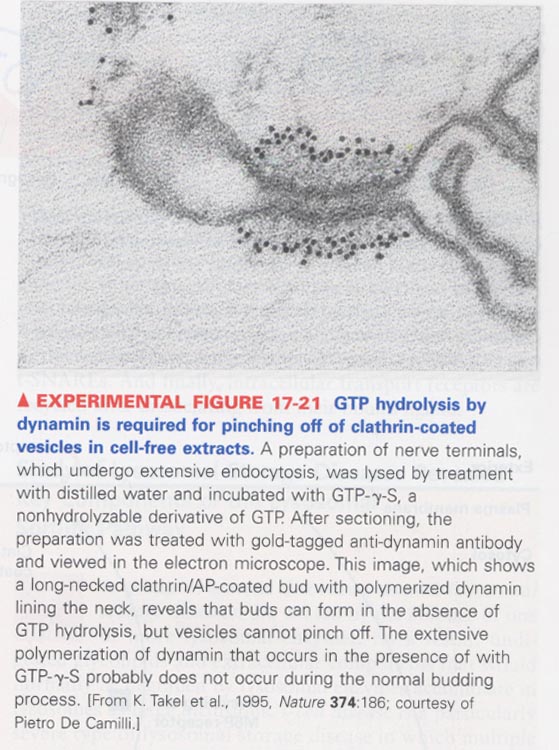
(8) Role of dynamin for pinching off of clathrin vesicles
--> GTP hydolysis; energy release and for contraction of the vesicle neck
--> in case of COPI and COPII vesicles, no-requirement of dynamin
evidence; use of a nonhydrolyzable GTP analog, GTP-γ-S and gold-tagged anti-dynamin antibody
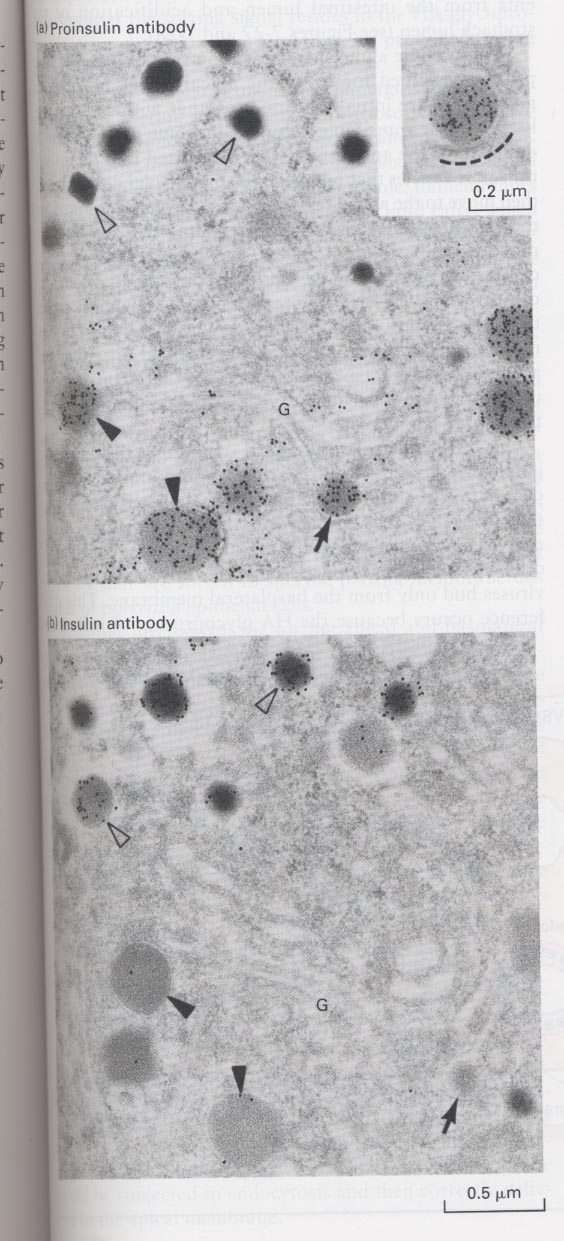
(9) Proteolytic cleavage of some membrane and secretory proteins after leaving the Trans-Golgi
kinds; some membrane and soluble secretory proteins
(ex) lysosomal enzymes, influenza hemagglutinin, albumin, insulin, glucagon, yeast α-mating factor
** in case of insulun
--> immature secretory vesicles (closed arrowheads) and vesicles budding from trans-Golgi (arrow)
--> contain only proproteins, not mature proteins
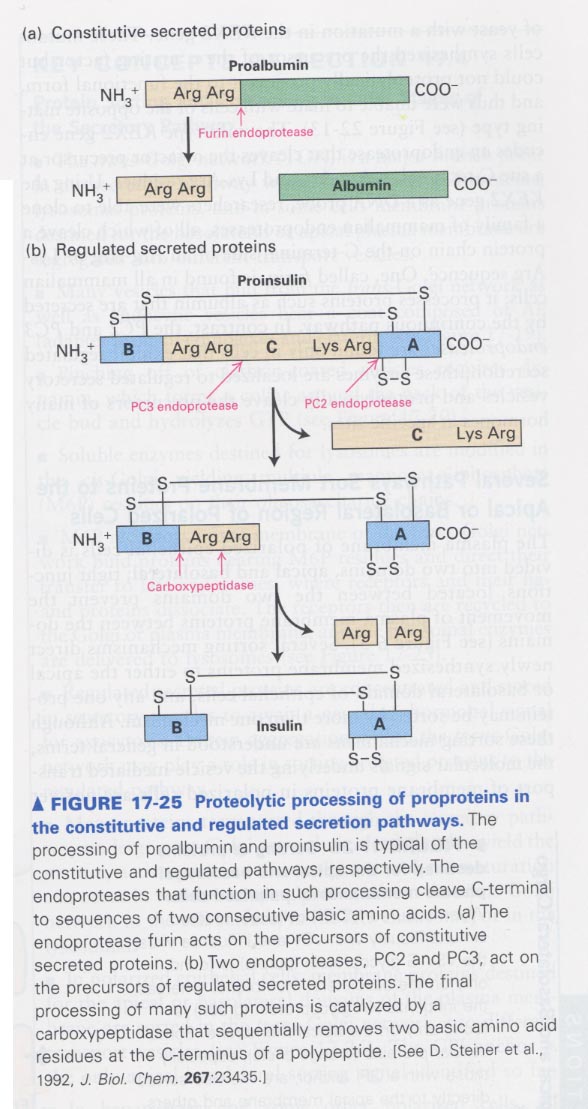
** proteolytic cleavage of albumin (a) and insulin (b)
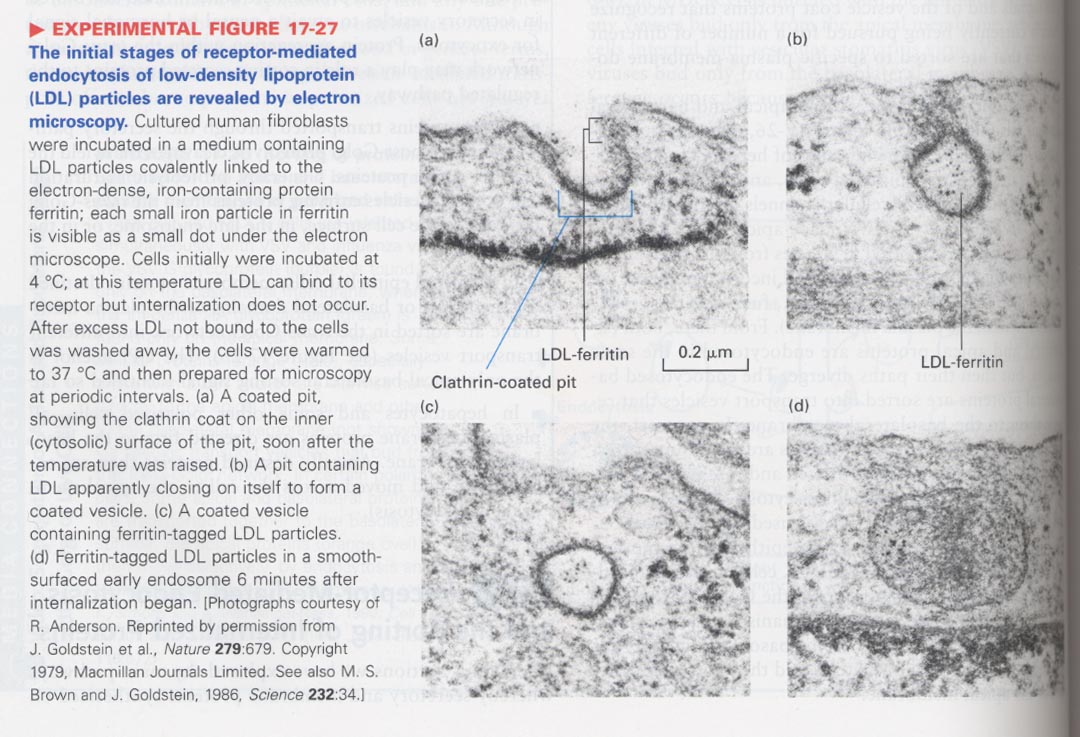
(10) Receptor-mediated endocytosis
--> LDL particle + ferritin
** Model for receptor-mediated endocytosis
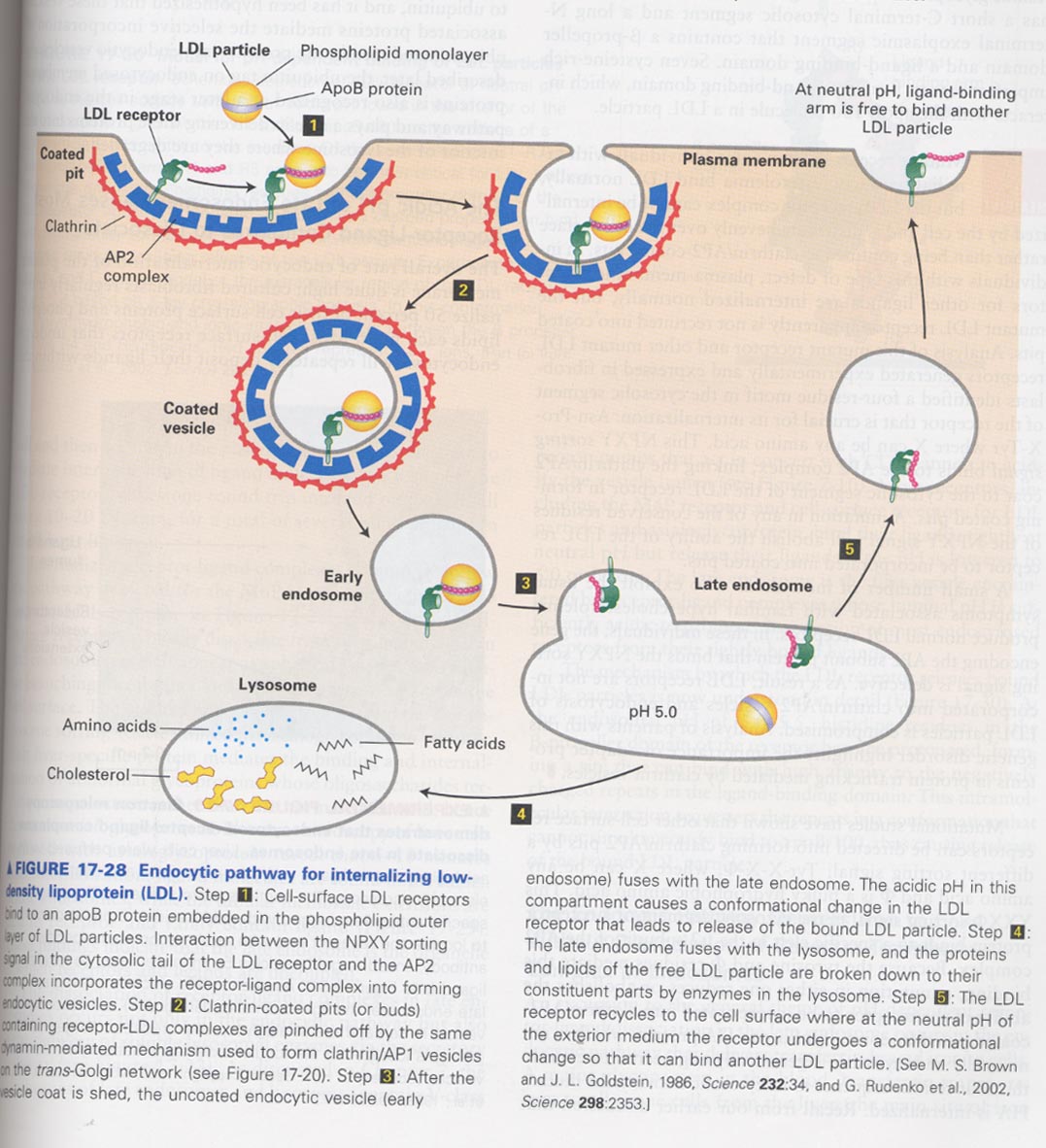
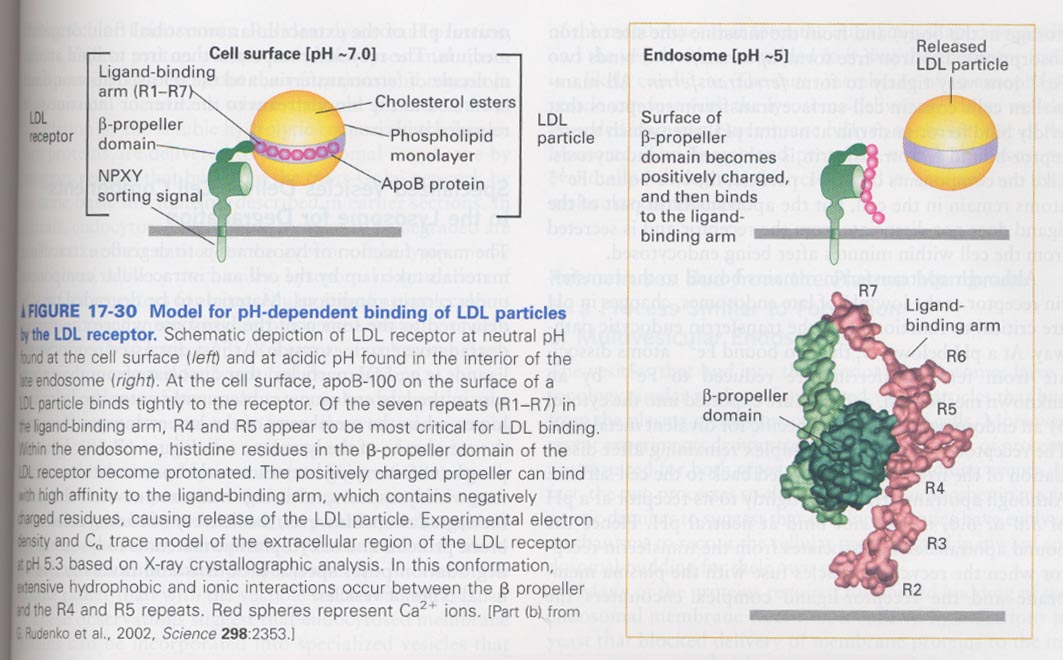
** binding model between LDL particle and LDL receptor
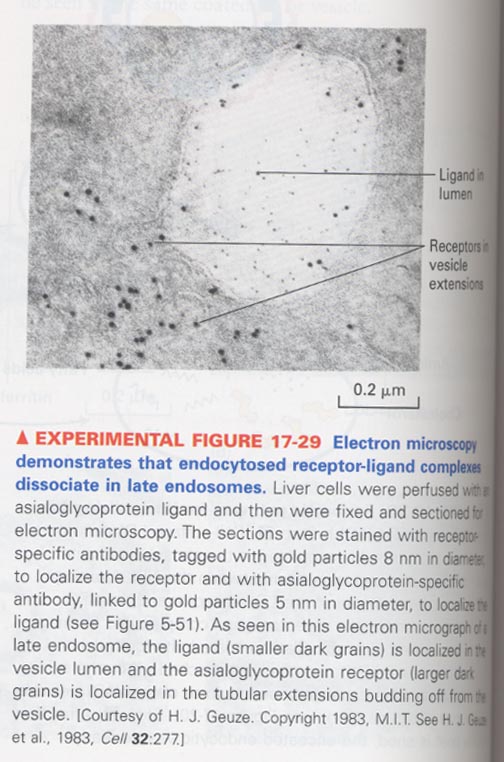
** dissociation of endocytosed receptor-ligand complexes
--> in late endosomes
--> asialoglycoprotein-specific antibody conjugated with gold particles
ex) LDL receptor; 1 turn/10-20 min
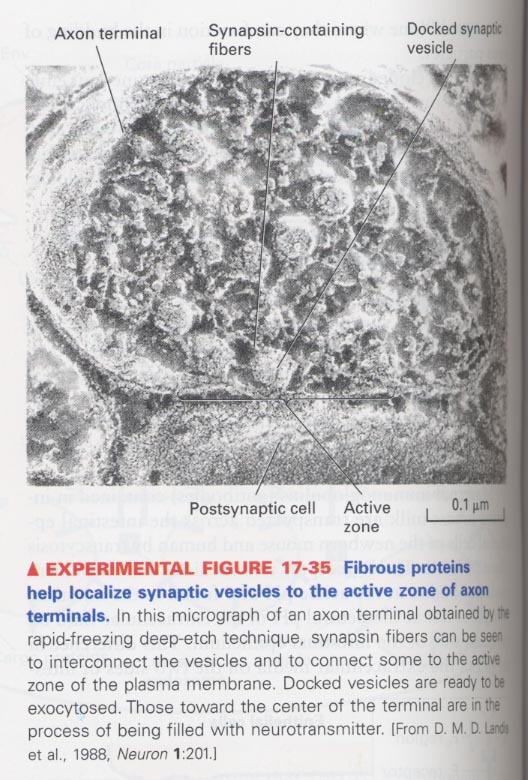
(11) Synaptic vesicle fusion and recycling
synaptic vesicles; located in active zone, contains a Ca2+-binding protein
** Model for synaptic vesicle recycling
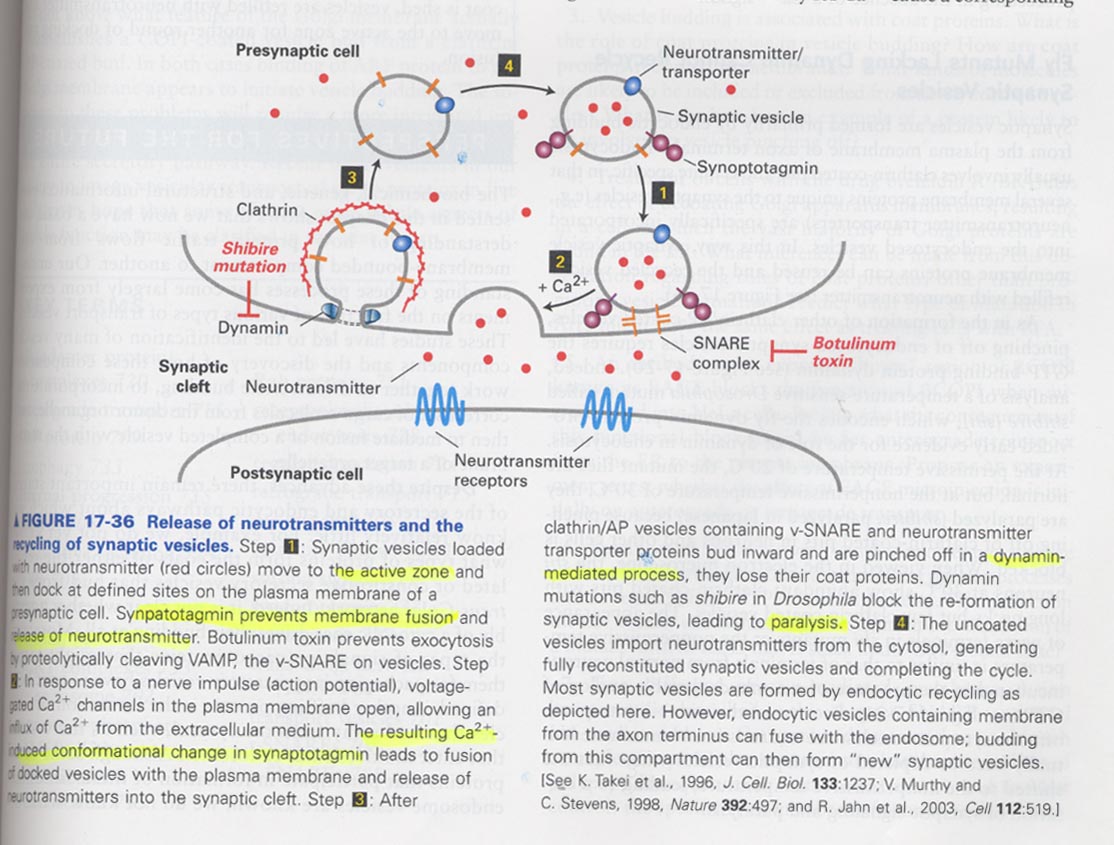
** synapsin-containg fibrous proteins help the localization of synaptic vesicles in active zones